BsiteP - predicts protein binding site
NuriKit - generates 3D general (non-macrocyclic) ligand conformations
GDDL - predicts protein-ligand binding poses When the protein and ligand engage in typical (non-covalent) interactions
BindingRMSD - predicts protein-ligand binding poses RMSD
#Section 0: Fetching and installing the required software
Please keep in mind that Google Colab requires a few minutes to initialize the hosted runtime at the beginning.
Before starting the toolkit, we need to download the necessary software. The types are as follows:
py3Dmol - helps to visualize protein or ligand structures
openbabel_wheel - helps interconversion of chemical file formats, molecular structure manipulation, and cheminformatics tasks.
rdkit - helps representing, manipulating, and analyzing chemical structures
prolif - helps analyze protein-ligand interactions
MDAnalysis - allows for efficient handling of the structures of the protein and ligand
torch_* - extends PyTorch’s capabilities for operations on tensors ( * includes geometric, scatter, cluster)
DGL - simplifies deep learning on graph-structured data, providing efficient tools for building graph neural networks
[ ]:
%%capture
! pip install py3Dmol
! pip install openbabel_wheel
! pip install rdkit
! pip install prolif
! pip install MDAnalysis
! pip install torch_geometric
! pip install dgl -f https://data.dgl.ai/wheels/torch-2.4/cu121/repo.html
Then, download the toolkit code stored in the GitHub repository
[ ]:
%%capture
# Install BsiteP
! git clone https://github.com/ding-oh/BsiteP
# Install BindingRMSD
! git clone https://github.com/eightmm/BindingRMSD
# Install NuriKit
! pip install nurikit
# Install GDDL
! git clone https://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@github.com/seoklab/colab_gd_dl.git
%cd /content/colab_gd_dl
! pip install -e .
Now, we are ready to use the toolkit.
#Section 1: Generating 3D conformations for two ligands using “nurikit”
1D SMILES for ligand 1 (drug name):
1D SMILES for ligand 2 (drug name):
#1-0. Choose whether the docking method is covalent or non-covalent#
If you select general (non-covalent) docking in this, proceed to 1-1.
If you select covalent docking, proceed to 1-2. This choice will also be reflected when selecting a docking tool later.
[ ]:
#@title ##Selecting docking mode (covalent or non-covalent)
whether_covalent_docking_or_not = "non-covalent" #@param ["covalent", "non-covalent"]
#@markdown - If the protein and ligand are connected by a covalent bond, select covalent; otherwise, non-covalent.
#1-1. For non-covalent docking#
If you are going to use covalent docking tool (C-Dock), Please skip this section and go to 1-2, please.
Please provide the SMILES format of the molecule you want to convert into a 3D conformation. NuriKit will quickly generate the ligand structure. If checking the show_ligand_3D_conformation, you can visually inspect the 3D structure of the molecule.
[ ]:
#@title Ligand input
import nuri.algo
from pathlib import Path
import nuri.algo
from openbabel import pybel
import py3Dmol
import os
from rdkit import Chem
from rdkit.Chem import AllChem
target_dir = f'/content/iitp_demonstrate/'
os.makedirs(target_dir, exist_ok=True)
%cd '/content/iitp_demonstrate//'
SMILES = "c1(c(cc2c(c1)c(ncn2)Nc1cc(c(cc1)F)Cl)OC)NC(=O)CCCN" #@param {type:"string"}
#@markdown - Please provide the SMILES format of the ligand to be used for protein-ligand docking
mol = next(nuri.readstring("smi", SMILES))
nuri.algo.generate_coords(mol)
Path("ligand_input.mol2").write_text(nuri.to_mol2(mol))
show_ligand_3D_conformation = True #@param {type:"boolean"}
/content/iitp_demonstrate
Hydrogens are added and charges are assigned to the generated structure.
[ ]:
mol = next(pybel.readfile("mol2", "ligand_input.mol2"))
mol.addh()
mol.calccharges("gasteiger")
mol.write("mol2", "ligand_charged.mol2", overwrite=True)
The unnecessary parts are removed from the ligand file.
[ ]:
wrt = []
write = True
with open('ligand_charged.mol2') as fp:
for line in fp:
if line == '@<TRIPOS>UNITY_ATOM_ATTR\n':
write = False
if line == '@<TRIPOS>BOND\n':
write = True
if write:
wrt.append(line)
fout= open('ligand_charged.mol2', 'wt')
fout.writelines(wrt)
fout.close()
If checking the show_ligand_3D_conformation above, you can visualize the 3D structure of the molecule.
[ ]:
#@title Displaying 3D structure
if show_ligand_3D_conformation:
print("")
print("\033[1m<Input ligand 3D conformation>\033[0m")
mol = next(pybel.readfile("mol2", "ligand_charged.mol2"))
pdb_data = mol.write("pdb")
view = py3Dmol.view(width=600, height=400)
view.addModel(pdb_data, "pdb")
view.setStyle({'stick': {}})
view.zoomTo()
view.show()
<Input ligand 3D conformation>
3Dmol.js failed to load for some reason. Please check your browser console for error messages.
You can download the file of generated ligand 3D conformation (mol2 format).
[ ]:
from google.colab import files
output_file = 'ligand_charged.mol2'
files.download(output_file)
Section 2: Finding binding pockets for a given protein (EGFR) using “BsiteP”¶
## Benchmarking results of binding site prediction method¶
We benchmarked our binding site prediction method on widely used binding site benchmark sets: COACH420 and BU48.
The success rate of binding site predictions are generally measured by two metrics: DCC and DCA.
DCC measures if the distance between the centers of mass of ligand and ligand binding site residues are less than a given cutoff distance.
DCA is measures if the distance between the any heavy atoms of a ligand and ligand binding site residues are less than a given cutoff distance.
Thus, DCA is easier to satify than DCC.
The plots below show that our method achived high accuracy in binding site predictions.
With the COACH420 benchmark set, our method achieved an accuracy of DCC > 65% and DCA > 90%.
With the BU48 benchmark set, our method achieved an accuracy of DCC > 70% and DCA > 93%.
[ ]:
#@title Setting up enviroments
%%capture
import sys
import os
if os.path.basename(os.getcwd()) != "BsiteP":
bsitep_path = "/content/BsiteP"
if os.path.exists(bsitep_path):
os.chdir(bsitep_path)
print(f"Changed directory to: {os.getcwd()}")
else:
print("Error: BsiteP directory does not exist in the current path.")
else:
print(f"Already in BsiteP directory: {os.getcwd()}")
import torch
from openbabel import pybel
from utils.model_utils import load_model
from utils.pocket_utils import save_pocket_mol
from utils.data_utils import prepare_data
import glob
from collections import OrderedDict
from SEResnet import SEResNet
# Check device availability
device = torch.device('cuda' if torch.cuda.is_available() else 'cpu')
model = SEResNet().to(device)
model_path = f"./ckpt/model0.pth"
state_dict = torch.load(model_path, map_location=device)
new_dict = OrderedDict((key[7:], value) for key, value in state_dict.items())
model.load_state_dict(new_dict)
model.to(device)
model.eval()
print(f"Model loaded successfully. - using device: {device}")
[ ]:
from google.colab import drive
drive.mount('/content/drive')
Mounted at /content/drive
[ ]:
import os
import requests
input_path = "./example"
output_path = "./output/example"
file_format = "pdb"
os.makedirs(input_path, exist_ok=True)
os.makedirs(output_path, exist_ok=True)
#@markdown - We present two options. The first is to download the pdb file from RCSB PDB and the second is to upload it to mydrive. Then, the prepared files are processed and placed in 'example'
#@markdown - Please provide the PDB ID or your PDB file.
Download_PDB = "4i24" # @param {type:"string"}
#@markdown - Download PDB file from RCSB PDB and process the downloaded pdb.
Uploaded_pdb_path = "/content/drive/MyDrive/your_pdb_file.pdb" # @param {type:"string"}
#@markdown - Please place your PDB file at '/content/drive/MyDrive/.
#@markdown - Process the uploaded pdb file from mydrive as '/example/{pdb_id}/protein.pdb'.
Target_chain = "ALL" # @param {type:"string"}
#@markdown - Target Chain (ex: A, B, C ...)
#@markdown - 시연 코드는 RCSB PDB로부터 2QEH을 받아서 사용함.
base_output_dir = "./example"
def find_pdb_id(pdb_lines):
for line in pdb_lines:
if line.startswith("HEADER"):
return line[62:66].strip() # PDB ID is typically at these positions in HEADER line
return None # Return None if no HEADER line with PDB ID is found
def process_pdb_file(pdb_lines, chain, output_dir):
protein_path = os.path.join(output_dir, "protein.pdb")
ligand_path = os.path.join(output_dir, "ligand.pdb")
with open(protein_path, 'w') as protein_file, open(ligand_path, 'w') as ligand_file:
for line in pdb_lines:
if line.startswith("ATOM"):
if chain.upper() == "ALL" or f" {chain} " in line:
protein_file.write(line + "\n") # Write ATOM lines for the selected chain
elif line.startswith("HETATM"):
if not (" DMS " in line or " HOH " in line):
ligand_file.write(line + "\n") # Write HETATM lines excluding DMS and HOH
print(protein_path)
print(ligand_path)
def handle_pdb_file(path, chain):
with open(path, 'r') as file:
pdb_lines = file.readlines()
pdb_id = find_pdb_id(pdb_lines) or "Unknown"
output_dir = os.path.join(base_output_dir, pdb_id)
os.makedirs(output_dir, exist_ok=True)
process_pdb_file(pdb_lines, chain, output_dir)
print(f"PDB processed and saved in directory: {output_dir}")
if Uploaded_pdb_path != "/content/drive/MyDrive/your_pdb_file.pdb":
if os.path.exists(Uploaded_pdb_path):
handle_pdb_file(Uploaded_pdb_path, Target_chain)
else:
print(f"Warning: The file at '{Uploaded_pdb_path}' does not exist. Please check the path and ensure the file is correctly placed.")
# Download and process from PDB ID if no uploaded file is processed
elif Download_PDB:
output_dir = os.path.join(base_output_dir, Download_PDB)
os.makedirs(output_dir, exist_ok=True)
protein_path = os.path.join(output_dir, "protein.pdb")
ligand_path = os.path.join(output_dir, "ligand.pdb")
print(f"Downloading PDB ID: {Download_PDB}")
url = f"https://files.rcsb.org/download/{Download_PDB}.pdb"
response = requests.get(url)
if response.status_code == 200:
pdb_lines = response.text.splitlines()
process_pdb_file(pdb_lines, Target_chain, output_dir)
print(f"PDB ID {Download_PDB} processed and saved in directory: {output_dir}")
else:
print(f"Failed to download PDB file {Download_PDB}. Status code: {response.status_code}")
else:
print("No valid PDB file provided or found.")
Downloading PDB ID: 4i24
./example/4i24/protein.pdb
./example/4i24/ligand.pdb
PDB ID 4i24 processed and saved in directory: ./example/4i24
[ ]:
#@title Visualizing predicted binding site with native ligand
%%capture
import os
from py3Dmol import view
def read_pdb_as_string(file_path):
"""Read the PDB file content as a string."""
with open(file_path, 'r') as file:
return file.read()
def visualize_molecules(protein_path, ligand_path=None, pocket_paths=None, label_residues=False):
"""
Visualizes the protein, ligand, and predicted pockets using Py3Dmol.
Adds residue labels with residue names and numbers for the pockets if `label_residues` is True.
"""
try:
viewer = view(width=1200, height=800)
# Add protein
protein_data = read_pdb_as_string(protein_path)
if protein_data:
viewer.addModel(protein_data, 'pdb')
viewer.setStyle({'model': 0}, {'cartoon': {'color': 'white'}})
# Add ligand
if ligand_path and os.path.exists(ligand_path):
ligand_data = read_pdb_as_string(ligand_path)
viewer.addModel(ligand_data, 'pdb')
viewer.setStyle({'model': 1}, {'stick': {'radius': 0.3, 'color': 'red'}})
# Add pockets and optionally label residues
if pocket_paths:
pocket_path = pocket_paths[0]
if os.path.exists(pocket_path):
pocket_data = read_pdb_as_string(pocket_path)
viewer.addModel(pocket_data, 'pdb')
viewer.setStyle({'model': 2 + i}, {'sphere': {'opacity': 0.4, 'color': 'blue'}})
if label_residues:
# Extract residues and label them
lines = pocket_data.splitlines()
for line in lines:
if line.startswith("ATOM") or line.startswith("HETATM"):
residue_name = line[17:20].strip()
residue_number = line[22:26].strip()
chain_id = line[21].strip()
viewer.addLabel(
f"{residue_name} {residue_number}",
{
'position': {
'x': float(line[30:38].strip()),
'y': float(line[38:46].strip()),
'z': float(line[46:54].strip())
},
'backgroundColor': 'white',
'fontColor': 'black',
'fontSize': 10
},
{'chain': chain_id, 'resi': int(residue_number)}
)
# Adjust view
viewer.zoomTo()
viewer.show()
except Exception as e:
print(f"An error occurred during visualization: {e}")
[ ]:
visualize_molecules(protein_path)
3Dmol.js failed to load for some reason. Please check your browser console for error messages.
[ ]:
#@title Preparing Input Data
%%capture
input_path = "./example"
output_path = "./output/example"
file_format = "pdb"
# Prepare input data
!rm -rf /content/BsiteP/example/.ipynb_checkpoints
input_data = prepare_data(input_path, file_format)
print(f"Prepared {len(input_data)} input samples.")
[ ]:
#@title Predicting Binding Pockets
%%capture
pocket_paths = []
for i, data in enumerate(input_data):
input_tensor, origin, step, name, mol_paths = data
mol = next(pybel.readfile(file_format, mol_paths[0]))
input_tensor = input_tensor.to(device)
# Perform inference
with torch.no_grad():
output = model(input_tensor)
# Save predicted pockets
pockets, score = save_pocket_mol(output.cpu(), origin[0], step[0], mol, dist_cutoff=1.5)
print(pockets)
# Save pocket files
for j, pocket in enumerate(pockets):
folder_name = os.path.join(output_path, str(name).split("'")[1])
os.makedirs(folder_name, exist_ok=True)
pocket_filename = os.path.join(folder_name, f"pocket{j}.{file_format}")
pocket.write(file_format, pocket_filename, overwrite=True)
pocket_paths.append(pocket_filename)
[ ]:
#@title Predicting Binding Pockets
import pandas as pd
AA_3TO1 = {
"ALA": "A", "ARG": "R", "ASN": "N", "ASP": "D", "CYS": "C",
"GLN": "Q", "GLU": "E", "GLY": "G", "HIS": "H", "ILE": "I",
"LEU": "L", "LYS": "K", "MET": "M", "PHE": "F", "PRO": "P",
"SER": "S", "THR": "T", "TRP": "W", "TYR": "Y", "VAL": "V"
}
def parse_coords(file):
coords = {}
try:
with open(file, 'r') as f:
for line in f:
if line.startswith("ATOM"):
res = line[17:20].strip()
chain = line[21].strip()
res_id = int(line[22:26].strip())
x, y, z = map(float, (line[30:38].strip(), line[38:46].strip(), line[46:54].strip()))
if res in AA_3TO1:
coords[(x, y, z)] = (res_id, chain, AA_3TO1[res])
except FileNotFoundError:
print("Failed : Model can not found any binding site.")
return coords
def match_residues(pocket_file, protein_coords):
pocket_coords = parse_coords(pocket_file)
return {protein_coords[pc][0] for pc in pocket_coords if pc in protein_coords}
def calculate_center(pocket_file):
coords = []
try:
with open(pocket_file, 'r') as f:
for line in f:
if line.startswith("ATOM"):
x, y, z = map(float, (line[30:38].strip(), line[38:46].strip(), line[46:54].strip()))
coords.append((x, y, z))
if coords:
x_mean = sum(c[0] for c in coords) / len(coords)
y_mean = sum(c[1] for c in coords) / len(coords)
z_mean = sum(c[2] for c in coords) / len(coords)
return x_mean, y_mean, z_mean
except FileNotFoundError:
pass
return None, None, None
def parse_sequence(file):
chains = {}
with open(file, 'r') as f:
for line in f:
if line.startswith("ATOM"):
res = line[17:20].strip()
chain = line[21].strip()
res_id = int(line[22:26].strip())
if res in AA_3TO1:
chains.setdefault(chain, {})[res_id] = AA_3TO1[res]
return chains
def generate_seqs(chains, matches):
full_seqs = {}
highlight_seqs = {}
for c, residues in chains.items():
full_seqs[c] = ''.join(residues[rid] for rid in sorted(residues))
highlight_seqs[c] = ''.join(residues[rid] if rid in matches else '-' for rid in sorted(residues))
return full_seqs, highlight_seqs
def format_split_seq(target_seq, binding_seq, interval=100):
result = []
for i in range(0, max(len(target_seq), len(binding_seq)), interval):
target_chunk = target_seq[i:i+interval]
binding_chunk = binding_seq[i:i+interval]
index_line = ''.join([f"{j:>5}" if (j+1) % interval == 0 else " " for j in range(i, i+len(target_chunk))])
result.append(f"{index_line}\n{target_chunk}\n{binding_chunk}")
return '\n\n'.join(result)
def print_pocket_table(chains, highlight_seqs):
pocket_data = {
"Chain": [],
"Residue ID": [],
"Amino Acid": []
}
for chain, seq in highlight_seqs.items():
for idx, residue in enumerate(seq, start=1):
if residue != '-':
pocket_data["Chain"].append(chain)
pocket_data["Residue ID"].append(idx)
pocket_data["Amino Acid"].append(residue)
df = pd.DataFrame(pocket_data)
if not df.empty:
print(df)
else:
print("No pocket residues found.")
protein_file = os.path.join(input_path, Download_PDB, "protein.pdb")
pocket_file = f'./output/example/{Download_PDB}/pocket0.pdb'
protein_coords = parse_coords(protein_file)
pocket_residues = match_residues(pocket_file, protein_coords)
chains = parse_sequence(protein_file)
full_seqs, highlight_seqs = generate_seqs(chains, pocket_residues)
center = calculate_center(pocket_file)
[ ]:
#@title Visualizing the protein, ligand, and pockets
ligand_file = os.path.join(input_path, Download_PDB, "ligand.pdb")
visualize_molecules(protein_path, ligand_file, pocket_paths, label_residues=False)
for chain in chains.keys():
print(f"Chain {chain}:")
print(format_split_seq(full_seqs[chain], highlight_seqs[chain]))
3Dmol.js failed to load for some reason. Please check your browser console for error messages.
Chain A:
99
NQALLRILKETEFKKIKVLGSGAFGTVYKGLWIPEGEKVKIPVAIKELANKEILDEAYVMASVDNPHVCRLLGICLTSTVQLIMQLMPFGCLLDYVREHK
------------------L-------V----------------A-K-----------------------------------L-M-LM--GC---------
199
DNIGSQYLLNWCVQIAKGMNYLEDRRLVHRDLAARNVLVKTPQHVKITDFGLAKLLVPIKWMALESILHRIYTHQSDVWSYGVTVWELMTFGSKPYDGIP
----------------------------------R--L---------TD---------------------------------------------------
ASEISSILEKGERLPQPPICTIDVYMIMVKCWMIDADSRPKFRELIIEFSKMARDPQRYLVIQGDERMHLPSPTDSNFYRALMDEEDMDDVVDAD
-----------------------------------------------------------------------------------------------
Chain B:
99
NQALLRILKETEFKKIKVLGSGAFGTVYKGLWIPEGEKVKIPVAIKELREATSPKANKEILDEAYVMASVDNPHVCRLLGICLTSTVQLIMQLMPFGCLL
------------------L-------V----------------A-K------------------------------------------L-M-LM--GC--
199
DYVREHKDNIGSQYLLNWCVQIAKGMNYLEDRRLVHRDLAARNVLVKTPQHVKITDFGLAKLLGKVPIKWMALESILHRIYTHQSDVWSYGVTVWELMTF
-----------------------------------------R--L---------TD--------------------------------------------
299
GSKPYDGIPASEISSILEKGERLPQPPICTIDVYMIMVKCWMIDADSRPKFRELIIEFSKMARDPQRYLVIQGDERMHLPSPTDSNFYRALMDEEDMDDV
----------------------------------------------------------------------------------------------------
VDAD
----
[ ]:
print_pocket_table(chains, highlight_seqs)
Chain Residue ID Amino Acid
0 A 19 L
1 A 27 V
2 A 44 A
3 A 46 K
4 A 82 L
5 A 84 M
6 A 86 L
7 A 87 M
8 A 90 G
9 A 91 C
10 A 135 R
11 A 138 L
12 A 148 T
13 A 149 D
14 B 19 L
15 B 27 V
16 B 44 A
17 B 46 K
18 B 89 L
19 B 91 M
20 B 93 L
21 B 94 M
22 B 97 G
23 B 98 C
24 B 142 R
25 B 145 L
26 B 155 T
27 B 156 D
#Section 3: Conducting Protein-ligand Docking Using GDDL¶
The accuracy of protein-ligand docking methods are evaluted with two popular benchmark sets: CASP-2016 and PoseBuster.
Generally, ligand pose prediction is considered accurate if RMSD value of a predicted pose is less than 2 angstrom from the crystal structure.
The plots below show the benchmark results of our protein-ligand docking program, GalaxyDock-DL (GDDL).
Using the CASF-2016 core set, GDDL showed a docking accuracy of 87%, which is comparable to existing deep-learning-based docking models.
Using the PoseBuster set, which is newer and more comprehensive than CASP-2016, GDDL showed a docking accuracy of 63%~78%, which is next to the best existing model, CarsiDock.
#3-0. Obtaining the binding site center from Section 2
Since both GDDL and C-Dock are local docking tools, the binding site center coordinates of the protein where the ligand will be docked are required. The binding site center information predicted in Section 2 is provided as input to the docking tools. f you want to specify the desired coordinates as the docking box center without using the binding site from Section 2, you can check the put_direct_coordinates option and enter the desired coordinates in the x, y, and z fields.
[ ]:
#@title Specifying the center coordinates of a ligand binding site
import os
import shutil
import subprocess
target = '4i24'
source_path = '/content/BsiteP/example/4i24/protein.pdb'
source_lig_path = '/content/BsiteP/example/4i24/ligand.pdb'
target_dir = '/content/iitp_demonstrate/'
target_file = os.path.join(target_dir, '4i24_pro.pdb')
lig_file = os.path.join(target_dir, '4i24_lig.pdb')
shutil.copy(source_path, target_file)
shutil.copy(source_lig_path, lig_file)
%cd /content/iitp_demonstrate/
put_direct_coordinates = True #@param{type:"boolean"}
if put_direct_coordinates:
x = '-12.57260799407959' #@param {type:"string"}
y = '41.4859619140625' #@param {type:"string"}
z = '-3.1989645957946777' #@param {type:"string"}
else:
x = center[0]
y = center[1]
z = center[2]
/content/iitp_demonstrate
If you select general (non-covalent docking) in section 1, proceed to 3-1. If you select non-covalent docking, proceed to 3-2.
#3-1. Protein-ligand docking 1: EGFR(L858R)-ligand1 using “GDDL” Protein-ligand docking 2: EGFR(L858R, T790M)-ligand1 using “galaxydock_dl” Protein-ligand docking 3: EGFR(L858R, T790M)-ligand2 using “galaxydock_dl”
GDDL can be run in two versions. The ‘original’ version uses the existing GDDL parameters, which increases the likelihood of obtaining more accurate results, but it takes longer (around 30 to 50 minutes). The ‘light’ version reduces the number of parameters to speed up the docking process (about 10 minutes), but the results may be relatively less accurate.
[ ]:
#@title Selecting gddl_docking version
gddl_docking_ver = "original" #@param ["original", "light"]
Protein-ligad docking by GDDL in assigned gddl_docking_ver is performed.
[ ]:
#@title Performing protein-ligand docking with GDDL
if whether_covalent_docking_or_not == 'non-covalent':
if gddl_docking_ver == 'original':
command = f'python /content/colab_gd_dl/scripts/run_gd_dl_from_other_directory.py -d /content/colab_gd_dl/ -p /content/iitp_demonstrate/{target}_pro.pdb -l /content/iitp_demonstrate/ligand_charged.mol2 --out_dir /content/iitp_demonstrate/ -x {x} -y {y} -z {z}'
process = subprocess.Popen(command, shell=True, stdout=subprocess.PIPE, stderr=subprocess.PIPE, text=True)
for line in process.stdout:
print(line, end='')
stderr_output = process.stderr.read()
if stderr_output:
print("Error:", stderr_output)
process.wait()
if gddl_docking_ver == 'light':
command = f'python /content/colab_gd_dl/scripts/run_gd_dl_from_other_directory.py -d /content/colab_gd_dl/ -p /content/iitp_demonstrate/{target}_pro.pdb -l /content/iitp_demonstrate/ligand_charged.mol2 --out_dir /content/iitp_demonstrate/ -x {x} -y {y} -z {z} --n_seed_cycle 1 --max_opt_cycle 10 --n_bank 50'
process = subprocess.Popen(command, shell=True, stdout=subprocess.PIPE, stderr=subprocess.PIPE, text=True)
for line in process.stdout:
print(line, end='')
stderr_output = process.stderr.read()
if stderr_output:
print("Error:", stderr_output)
process.wait()
스트리밍 출력 내용이 길어서 마지막 5000줄이 삭제되었습니다.
------------------------------------------------------------
Select Seeds....
Unused seeds : 11 / 100
------------------------------------------------------------
Make new conformation
perturbation rate : 0.000
calc_ml_energy.mol2 is working!
Time for inference: 42.82 (s)
61 7.111 was replaced to 84 6.941 in same group
43 0.267 was replaced to 97 -0.014 in same group
32 5.466 was replaced to 132 5.182 in same group
12 5.543 was replaced to 178 4.074 in same group
24 7.128 was replaced to 182 7.059 in same group
24 7.059 was replaced to 184 4.640 in same group
39 6.825 was replaced to 190 6.449 in same group
76 5.429 was replaced to 199 5.386 in same group
61 6.941 was replaced to 210 5.223 in same group
Bank No.: Energy
Bank 1: 7.902
Bank 2: -8.143
Bank 3: -11.490
Bank 4: -15.768
Bank 5: -17.503
Bank 6: -12.428
Bank 7: -16.096
Bank 8: -15.499
Bank 9: -6.440
Bank 10: -18.253
Bank 11: -10.754
Bank 12: -14.252
Bank 13: -12.578
Bank 14: -17.096
Bank 15: -3.817
Bank 16: 18.151
Bank 17: -12.520
Bank 18: -12.484
Bank 19: -17.973
Bank 20: -1.594
Bank 21: -9.337
Bank 22: 8.983
Bank 23: -16.139
Bank 24: -3.031
Bank 25: 2.476
Bank 26: -12.710
Bank 27: 1.413
Bank 28: -13.952
Bank 29: -14.235
Bank 30: -1.944
Bank 31: -13.349
Bank 32: -0.261
Bank 33: -17.383
Bank 34: -13.259
Bank 35: -16.676
Bank 36: -17.961
Bank 37: -1.174
Bank 38: -16.756
Bank 39: -13.855
Bank 40: -16.890
Bank 41: -15.273
Bank 42: -10.572
Bank 43: -14.115
Bank 44: -7.125
Bank 45: -15.998
Bank 46: -2.847
Bank 47: -11.080
Bank 48: -18.291
Bank 49: -3.911
Bank 50: -10.887
Bank 51: -7.725
Bank 52: -6.062
Bank 53: -8.530
Bank 54: -17.361
Bank 55: -10.692
Bank 56: -13.383
Bank 57: -17.716
Bank 58: -12.285
Bank 59: -10.040
Bank 60: -9.144
Bank 61: -13.903
Bank 62: -9.283
Bank 63: -14.577
Bank 64: -17.229
Bank 65: -15.745
Bank 66: -12.305
Bank 67: -12.252
Bank 68: -9.073
Bank 69: -12.832
Bank 70: -15.190
Bank 71: -4.663
Bank 72: -19.086
Bank 73: -18.355
Bank 74: -11.687
Bank 75: -9.666
Bank 76: -14.137
Bank 77: -15.629
Bank 78: -4.390
Bank 79: -17.076
Bank 80: -13.944
Bank 81: -15.962
Bank 82: -11.243
Bank 83: -13.821
Bank 84: -16.847
Bank 85: -5.422
Bank 86: -13.892
Bank 87: -11.549
Bank 88: -16.065
Bank 89: -17.945
Bank 90: -9.604
Bank 91: -20.447
Bank 92: -15.901
Bank 93: 5.350
Bank 94: -12.538
Bank 95: -18.945
Bank 96: -19.477
Bank 97: -18.993
Bank 98: -12.528
Bank 99: -10.422
Bank 100: -15.040
------------------------------------------------------------
CSA Run Number / Interation / SEED CYCLE = 1 / 1 / 55
D_ave / D_cut / D_min = 5.674 / 2.078 / 2.078
------------------------------------------------------------
Used time for CSA cycle 0D 0H 44M 53.8S
LCSA PLD: 1 0 13600 2.078 54 52 -17.361 -6.062 3346473 7 100 2693.797 sec
------------------------------------------------------------
Select Seeds....
Unused seeds : 7 / 100
------------------------------------------------------------
Make new conformation
perturbation rate : 0.000
calc_ml_energy.mol2 is working!
Time for inference: 40.93 (s)
52 7.248 was replaced to 1 6.962 in new group
29 5.717 was replaced to 50 5.693 in same group
16 6.861 was replaced to 56 3.978 in same group
76 5.386 was replaced to 78 4.729 in same group
38 6.693 was replaced to 83 6.571 in same group
31 6.565 was replaced to 125 6.085 in same group
12 4.074 was replaced to 126 3.675 in same group
24 4.640 was replaced to 128 4.021 in same group
1 7.072 was replaced to 195 5.223 in new group
97 -2.775 was replaced to 202 -3.055 in same group
100 4.221 was replaced to 218 4.181 in same group
100 4.181 was replaced to 220 4.078 in same group
86 6.546 was replaced to 237 5.953 in same group
Bank No.: Energy
Bank 1: -6.232
Bank 2: -8.143
Bank 3: -11.490
Bank 4: -15.768
Bank 5: -17.503
Bank 6: -12.428
Bank 7: -16.096
Bank 8: -15.499
Bank 9: -6.440
Bank 10: -18.253
Bank 11: -10.754
Bank 12: -14.395
Bank 13: -12.578
Bank 14: -17.096
Bank 15: -3.817
Bank 16: 0.329
Bank 17: -12.520
Bank 18: -12.484
Bank 19: -17.973
Bank 20: -1.594
Bank 21: -9.337
Bank 22: 8.983
Bank 23: -16.139
Bank 24: -5.520
Bank 25: 2.476
Bank 26: -12.710
Bank 27: 1.413
Bank 28: -13.952
Bank 29: -15.404
Bank 30: -1.944
Bank 31: -12.930
Bank 32: -0.261
Bank 33: -17.383
Bank 34: -13.259
Bank 35: -16.676
Bank 36: -17.961
Bank 37: -1.174
Bank 38: -16.448
Bank 39: -13.855
Bank 40: -16.890
Bank 41: -15.273
Bank 42: -10.572
Bank 43: -14.115
Bank 44: -7.125
Bank 45: -15.998
Bank 46: -2.847
Bank 47: -11.080
Bank 48: -18.291
Bank 49: -3.911
Bank 50: -10.887
Bank 51: -7.725
Bank 52: -12.343
Bank 53: -8.530
Bank 54: -17.361
Bank 55: -10.692
Bank 56: -13.383
Bank 57: -17.716
Bank 58: -12.285
Bank 59: -10.040
Bank 60: -9.144
Bank 61: -13.903
Bank 62: -9.283
Bank 63: -14.577
Bank 64: -17.229
Bank 65: -15.745
Bank 66: -12.305
Bank 67: -12.252
Bank 68: -9.073
Bank 69: -12.832
Bank 70: -15.190
Bank 71: -4.663
Bank 72: -19.086
Bank 73: -18.355
Bank 74: -11.687
Bank 75: -9.666
Bank 76: -8.720
Bank 77: -15.629
Bank 78: -4.390
Bank 79: -17.076
Bank 80: -13.944
Bank 81: -15.962
Bank 82: -11.243
Bank 83: -13.821
Bank 84: -16.847
Bank 85: -5.422
Bank 86: -12.478
Bank 87: -11.549
Bank 88: -16.065
Bank 89: -17.945
Bank 90: -9.604
Bank 91: -20.447
Bank 92: -15.901
Bank 93: 5.350
Bank 94: -12.538
Bank 95: -18.945
Bank 96: -19.477
Bank 97: -18.377
Bank 98: -12.528
Bank 99: -10.422
Bank 100: -11.007
------------------------------------------------------------
CSA Run Number / Interation / SEED CYCLE = 1 / 1 / 56
D_ave / D_cut / D_min = 5.664 / 2.078 / 2.078
------------------------------------------------------------
Used time for CSA cycle 0D 0H 45M 43.5S
LCSA PLD: 1 0 13850 2.078 54 50 -17.361 -10.887 3404729 12 100 2743.493 sec
------------------------------------------------------------
Select Seeds....
Unused seeds : 12 / 100
------------------------------------------------------------
Make new conformation
perturbation rate : 0.000
calc_ml_energy.mol2 is working!
Time for inference: 41.58 (s)
12 3.675 was replaced to 54 3.385 in same group
50 7.067 was replaced to 56 5.442 in new group
24 4.021 was replaced to 133 3.881 in same group
86 5.953 was replaced to 205 5.599 in same group
5 -1.474 was replaced to 206 -2.193 in same group
91 -0.240 was replaced to 212 -1.661 in same group
84 3.857 was replaced to 224 3.818 in same group
43 -0.014 was replaced to 243 -0.794 in same group
Bank No.: Energy
Bank 1: -6.232
Bank 2: -8.143
Bank 3: -11.490
Bank 4: -15.768
Bank 5: -15.025
Bank 6: -12.428
Bank 7: -16.096
Bank 8: -15.499
Bank 9: -6.440
Bank 10: -18.253
Bank 11: -10.754
Bank 12: -14.173
Bank 13: -12.578
Bank 14: -17.096
Bank 15: -3.817
Bank 16: 0.329
Bank 17: -12.520
Bank 18: -12.484
Bank 19: -17.973
Bank 20: -1.594
Bank 21: -9.337
Bank 22: 8.983
Bank 23: -16.139
Bank 24: -6.850
Bank 25: 2.476
Bank 26: -12.710
Bank 27: 1.413
Bank 28: -13.952
Bank 29: -15.404
Bank 30: -1.944
Bank 31: -12.930
Bank 32: -0.261
Bank 33: -17.383
Bank 34: -13.259
Bank 35: -16.676
Bank 36: -17.961
Bank 37: -1.174
Bank 38: -16.448
Bank 39: -13.855
Bank 40: -16.890
Bank 41: -15.273
Bank 42: -10.572
Bank 43: -7.233
Bank 44: -7.125
Bank 45: -15.998
Bank 46: -2.847
Bank 47: -11.080
Bank 48: -18.291
Bank 49: -3.911
Bank 50: -12.721
Bank 51: -7.725
Bank 52: -12.343
Bank 53: -8.530
Bank 54: -17.361
Bank 55: -10.692
Bank 56: -13.383
Bank 57: -17.716
Bank 58: -12.285
Bank 59: -10.040
Bank 60: -9.144
Bank 61: -13.903
Bank 62: -9.283
Bank 63: -14.577
Bank 64: -17.229
Bank 65: -15.745
Bank 66: -12.305
Bank 67: -12.252
Bank 68: -9.073
Bank 69: -12.832
Bank 70: -15.190
Bank 71: -4.663
Bank 72: -19.086
Bank 73: -18.355
Bank 74: -11.687
Bank 75: -9.666
Bank 76: -8.720
Bank 77: -15.629
Bank 78: -4.390
Bank 79: -17.076
Bank 80: -13.944
Bank 81: -15.962
Bank 82: -11.243
Bank 83: -13.821
Bank 84: -17.026
Bank 85: -5.422
Bank 86: -12.399
Bank 87: -11.549
Bank 88: -16.065
Bank 89: -17.945
Bank 90: -9.604
Bank 91: -19.070
Bank 92: -15.901
Bank 93: 5.350
Bank 94: -12.538
Bank 95: -18.945
Bank 96: -19.477
Bank 97: -18.377
Bank 98: -12.528
Bank 99: -10.422
Bank 100: -11.007
------------------------------------------------------------
CSA Run Number / Interation / SEED CYCLE = 1 / 1 / 57
D_ave / D_cut / D_min = 5.666 / 2.078 / 2.078
------------------------------------------------------------
Used time for CSA cycle 0D 0H 46M 34.0S
LCSA PLD: 1 0 14100 2.078 54 13 -17.361 -12.578 3462822 8 100 2794.034 sec
------------------------------------------------------------
Select Seeds....
Unused seeds : 8 / 100
------------------------------------------------------------
Make new conformation
perturbation rate : 0.000
calc_ml_energy.mol2 is working!
Time for inference: 40.43 (s)
10 3.307 was replaced to 60 2.947 in same group
26 3.840 was replaced to 125 1.834 in same group
5 -2.193 was replaced to 126 -2.232 in same group
50 5.442 was replaced to 135 4.718 in same group
64 1.580 was replaced to 171 1.325 in same group
38 6.571 was replaced to 204 6.083 in same group
19 0.220 was replaced to 235 -0.125 in same group
Bank No.: Energy
Bank 1: -6.232
Bank 2: -8.143
Bank 3: -11.490
Bank 4: -15.768
Bank 5: -17.438
Bank 6: -12.428
Bank 7: -16.096
Bank 8: -15.499
Bank 9: -6.440
Bank 10: -6.531
Bank 11: -10.754
Bank 12: -14.173
Bank 13: -12.578
Bank 14: -17.096
Bank 15: -3.817
Bank 16: 0.329
Bank 17: -12.520
Bank 18: -12.484
Bank 19: -18.745
Bank 20: -1.594
Bank 21: -9.337
Bank 22: 8.983
Bank 23: -16.139
Bank 24: -6.850
Bank 25: 2.476
Bank 26: -16.850
Bank 27: 1.413
Bank 28: -13.952
Bank 29: -15.404
Bank 30: -1.944
Bank 31: -12.930
Bank 32: -0.261
Bank 33: -17.383
Bank 34: -13.259
Bank 35: -16.676
Bank 36: -17.961
Bank 37: -1.174
Bank 38: -14.482
Bank 39: -13.855
Bank 40: -16.890
Bank 41: -15.273
Bank 42: -10.572
Bank 43: -7.233
Bank 44: -7.125
Bank 45: -15.998
Bank 46: -2.847
Bank 47: -11.080
Bank 48: -18.291
Bank 49: -3.911
Bank 50: 9.560
Bank 51: -7.725
Bank 52: -12.343
Bank 53: -8.530
Bank 54: -17.361
Bank 55: -10.692
Bank 56: -13.383
Bank 57: -17.716
Bank 58: -12.285
Bank 59: -10.040
Bank 60: -9.144
Bank 61: -13.903
Bank 62: -9.283
Bank 63: -14.577
Bank 64: -16.476
Bank 65: -15.745
Bank 66: -12.305
Bank 67: -12.252
Bank 68: -9.073
Bank 69: -12.832
Bank 70: -15.190
Bank 71: -4.663
Bank 72: -19.086
Bank 73: -18.355
Bank 74: -11.687
Bank 75: -9.666
Bank 76: -8.720
Bank 77: -15.629
Bank 78: -4.390
Bank 79: -17.076
Bank 80: -13.944
Bank 81: -15.962
Bank 82: -11.243
Bank 83: -13.821
Bank 84: -17.026
Bank 85: -5.422
Bank 86: -12.399
Bank 87: -11.549
Bank 88: -16.065
Bank 89: -17.945
Bank 90: -9.604
Bank 91: -19.070
Bank 92: -15.901
Bank 93: 5.350
Bank 94: -12.538
Bank 95: -18.945
Bank 96: -19.477
Bank 97: -18.377
Bank 98: -12.528
Bank 99: -10.422
Bank 100: -11.007
------------------------------------------------------------
CSA Run Number / Interation / SEED CYCLE = 1 / 1 / 58
D_ave / D_cut / D_min = 5.669 / 2.078 / 2.078
------------------------------------------------------------
Used time for CSA cycle 0D 0H 47M 23.9S
LCSA PLD: 1 0 14350 2.078 54 13 -17.361 -12.578 3527691 7 100 2843.876 sec
------------------------------------------------------------
Select Seeds....
Unused seeds : 7 / 100
------------------------------------------------------------
Make new conformation
perturbation rate : 0.000
calc_ml_energy.mol2 is working!
Time for inference: 39.08 (s)
13 7.054 was replaced to 2 5.150 in new group
19 -0.125 was replaced to 51 -0.467 in same group
13 5.150 was replaced to 53 3.674 in same group
40 0.822 was replaced to 66 0.543 in same group
40 0.543 was replaced to 114 0.337 in same group
65 -1.147 was replaced to 169 -1.750 in same group
92 -0.676 was replaced to 178 -3.162 in same group
19 -0.467 was replaced to 182 -0.474 in same group
26 1.834 was replaced to 185 1.137 in same group
64 1.325 was replaced to 186 1.149 in same group
61 5.223 was replaced to 200 5.211 in same group
Bank No.: Energy
Bank 1: -6.232
Bank 2: -8.143
Bank 3: -11.490
Bank 4: -15.768
Bank 5: -17.438
Bank 6: -12.428
Bank 7: -16.096
Bank 8: -15.499
Bank 9: -6.440
Bank 10: -6.531
Bank 11: -10.754
Bank 12: -14.173
Bank 13: -13.727
Bank 14: -17.096
Bank 15: -3.817
Bank 16: 0.329
Bank 17: -12.520
Bank 18: -12.484
Bank 19: -18.843
Bank 20: -1.594
Bank 21: -9.337
Bank 22: 8.983
Bank 23: -16.139
Bank 24: -6.850
Bank 25: 2.476
Bank 26: -18.109
Bank 27: 1.413
Bank 28: -13.952
Bank 29: -15.404
Bank 30: -1.944
Bank 31: -12.930
Bank 32: -0.261
Bank 33: -17.383
Bank 34: -13.259
Bank 35: -16.676
Bank 36: -17.961
Bank 37: -1.174
Bank 38: -14.482
Bank 39: -13.855
Bank 40: -14.978
Bank 41: -15.273
Bank 42: -10.572
Bank 43: -7.233
Bank 44: -7.125
Bank 45: -15.998
Bank 46: -2.847
Bank 47: -11.080
Bank 48: -18.291
Bank 49: -3.911
Bank 50: 9.560
Bank 51: -7.725
Bank 52: -12.343
Bank 53: -8.530
Bank 54: -17.361
Bank 55: -10.692
Bank 56: -13.383
Bank 57: -17.716
Bank 58: -12.285
Bank 59: -10.040
Bank 60: -9.144
Bank 61: -17.464
Bank 62: -9.283
Bank 63: -14.577
Bank 64: -17.312
Bank 65: -19.457
Bank 66: -12.305
Bank 67: -12.252
Bank 68: -9.073
Bank 69: -12.832
Bank 70: -15.190
Bank 71: -4.663
Bank 72: -19.086
Bank 73: -18.355
Bank 74: -11.687
Bank 75: -9.666
Bank 76: -8.720
Bank 77: -15.629
Bank 78: -4.390
Bank 79: -17.076
Bank 80: -13.944
Bank 81: -15.962
Bank 82: -11.243
Bank 83: -13.821
Bank 84: -17.026
Bank 85: -5.422
Bank 86: -12.399
Bank 87: -11.549
Bank 88: -16.065
Bank 89: -17.945
Bank 90: -9.604
Bank 91: -19.070
Bank 92: -15.274
Bank 93: 5.350
Bank 94: -12.538
Bank 95: -18.945
Bank 96: -19.477
Bank 97: -18.377
Bank 98: -12.528
Bank 99: -10.422
Bank 100: -11.007
------------------------------------------------------------
CSA Run Number / Interation / SEED CYCLE = 1 / 1 / 59
D_ave / D_cut / D_min = 5.662 / 2.078 / 2.078
------------------------------------------------------------
Used time for CSA cycle 0D 0H 48M 12.2S
LCSA PLD: 1 0 14600 2.078 54 21 -17.361 -9.337 3583215 8 100 2892.187 sec
------------------------------------------------------------
Select Seeds....
Unused seeds : 8 / 100
------------------------------------------------------------
Make new conformation
perturbation rate : 0.000
calc_ml_energy.mol2 is working!
Time for inference: 41.01 (s)
40 0.337 was replaced to 132 -0.291 in same group
65 -1.750 was replaced to 138 -1.920 in same group
34 4.679 was replaced to 171 4.402 in same group
94 1.812 was replaced to 175 1.681 in same group
13 3.674 was replaced to 177 3.503 in same group
65 -1.920 was replaced to 195 -4.409 in same group
77 -1.619 was replaced to 206 -2.209 in same group
77 -2.209 was replaced to 208 -3.084 in same group
39 6.449 was replaced to 229 6.242 in same group
Bank No.: Energy
Bank 1: -6.232
Bank 2: -8.143
Bank 3: -11.490
Bank 4: -15.768
Bank 5: -17.438
Bank 6: -12.428
Bank 7: -16.096
Bank 8: -15.499
Bank 9: -6.440
Bank 10: -6.531
Bank 11: -10.754
Bank 12: -14.173
Bank 13: -14.324
Bank 14: -17.096
Bank 15: -3.817
Bank 16: 0.329
Bank 17: -12.520
Bank 18: -12.484
Bank 19: -18.843
Bank 20: -1.594
Bank 21: -9.337
Bank 22: 8.983
Bank 23: -16.139
Bank 24: -6.850
Bank 25: 2.476
Bank 26: -18.109
Bank 27: 1.413
Bank 28: -13.952
Bank 29: -15.404
Bank 30: -1.944
Bank 31: -12.930
Bank 32: -0.261
Bank 33: -17.383
Bank 34: -16.483
Bank 35: -16.676
Bank 36: -17.961
Bank 37: -1.174
Bank 38: -14.482
Bank 39: -10.748
Bank 40: -15.799
Bank 41: -15.273
Bank 42: -10.572
Bank 43: -7.233
Bank 44: -7.125
Bank 45: -15.998
Bank 46: -2.847
Bank 47: -11.080
Bank 48: -18.291
Bank 49: -3.911
Bank 50: 9.560
Bank 51: -7.725
Bank 52: -12.343
Bank 53: -8.530
Bank 54: -17.361
Bank 55: -10.692
Bank 56: -13.383
Bank 57: -17.716
Bank 58: -12.285
Bank 59: -10.040
Bank 60: -9.144
Bank 61: -17.464
Bank 62: -9.283
Bank 63: -14.577
Bank 64: -17.312
Bank 65: -19.571
Bank 66: -12.305
Bank 67: -12.252
Bank 68: -9.073
Bank 69: -12.832
Bank 70: -15.190
Bank 71: -4.663
Bank 72: -19.086
Bank 73: -18.355
Bank 74: -11.687
Bank 75: -9.666
Bank 76: -8.720
Bank 77: -16.726
Bank 78: -4.390
Bank 79: -17.076
Bank 80: -13.944
Bank 81: -15.962
Bank 82: -11.243
Bank 83: -13.821
Bank 84: -17.026
Bank 85: -5.422
Bank 86: -12.399
Bank 87: -11.549
Bank 88: -16.065
Bank 89: -17.945
Bank 90: -9.604
Bank 91: -19.070
Bank 92: -15.274
Bank 93: 5.350
Bank 94: 39.963
Bank 95: -18.945
Bank 96: -19.477
Bank 97: -18.377
Bank 98: -12.528
Bank 99: -10.422
Bank 100: -11.007
------------------------------------------------------------
CSA Run Number / Interation / SEED CYCLE = 1 / 1 / 60
D_ave / D_cut / D_min = 5.667 / 2.078 / 2.078
------------------------------------------------------------
Used time for CSA cycle 0D 0H 49M 2.1S
LCSA PLD: 1 0 14850 2.078 65 21 -19.571 -9.337 3644781 7 100 2942.142 sec
------------------------------------------------------------
Select Seeds....
Unused seeds : 7 / 100
------------------------------------------------------------
Make new conformation
perturbation rate : 0.000
calc_ml_energy.mol2 is working!
Time for inference: 39.75 (s)
7 0.747 was replaced to 88 -0.089 in same group
54 -3.860 was replaced to 110 -4.336 in same group
94 1.681 was replaced to 194 0.039 in same group
94 0.039 was replaced to 196 -0.361 in same group
61 5.211 was replaced to 199 5.027 in same group
81 1.682 was replaced to 213 -0.292 in same group
Bank No.: Energy
Bank 1: -6.232
Bank 2: -8.143
Bank 3: -11.490
Bank 4: -15.768
Bank 5: -17.438
Bank 6: -12.428
Bank 7: -16.853
Bank 8: -15.499
Bank 9: -6.440
Bank 10: -6.531
Bank 11: -10.754
Bank 12: -14.173
Bank 13: -14.324
Bank 14: -17.096
Bank 15: -3.817
Bank 16: 0.329
Bank 17: -12.520
Bank 18: -12.484
Bank 19: -18.843
Bank 20: -1.594
Bank 21: -9.337
Bank 22: 8.983
Bank 23: -16.139
Bank 24: -6.850
Bank 25: 2.476
Bank 26: -18.109
Bank 27: 1.413
Bank 28: -13.952
Bank 29: -15.404
Bank 30: -1.944
Bank 31: -12.930
Bank 32: -0.261
Bank 33: -17.383
Bank 34: -16.483
Bank 35: -16.676
Bank 36: -17.961
Bank 37: -1.174
Bank 38: -14.482
Bank 39: -10.748
Bank 40: -15.799
Bank 41: -15.273
Bank 42: -10.572
Bank 43: -7.233
Bank 44: -7.125
Bank 45: -15.998
Bank 46: -2.847
Bank 47: -11.080
Bank 48: -18.291
Bank 49: -3.911
Bank 50: 9.560
Bank 51: -7.725
Bank 52: -12.343
Bank 53: -8.530
Bank 54: -18.240
Bank 55: -10.692
Bank 56: -13.383
Bank 57: -17.716
Bank 58: -12.285
Bank 59: -10.040
Bank 60: -9.144
Bank 61: -16.307
Bank 62: -9.283
Bank 63: -14.577
Bank 64: -17.312
Bank 65: -19.571
Bank 66: -12.305
Bank 67: -12.252
Bank 68: -9.073
Bank 69: -12.832
Bank 70: -15.190
Bank 71: -4.663
Bank 72: -19.086
Bank 73: -18.355
Bank 74: -11.687
Bank 75: -9.666
Bank 76: -8.720
Bank 77: -16.726
Bank 78: -4.390
Bank 79: -17.076
Bank 80: -13.944
Bank 81: -10.018
Bank 82: -11.243
Bank 83: -13.821
Bank 84: -17.026
Bank 85: -5.422
Bank 86: -12.399
Bank 87: -11.549
Bank 88: -16.065
Bank 89: -17.945
Bank 90: -9.604
Bank 91: -19.070
Bank 92: -15.274
Bank 93: 5.350
Bank 94: -12.657
Bank 95: -18.945
Bank 96: -19.477
Bank 97: -18.377
Bank 98: -12.528
Bank 99: -10.422
Bank 100: -11.007
------------------------------------------------------------
CSA Run Number / Interation / SEED CYCLE = 1 / 1 / 61
D_ave / D_cut / D_min = 5.669 / 2.078 / 2.078
------------------------------------------------------------
Used time for CSA cycle 0D 0H 49M 51.3S
LCSA PLD: 1 0 15100 2.078 65 21 -19.571 -9.337 3706798 5 100 2991.258 sec
------------------------------------------------------------
Select Seeds....
Unused seeds : 5 / 100
------------------------------------------------------------
Make new conformation
perturbation rate : 0.000
calc_ml_energy.mol2 is working!
Time for inference: 41.39 (s)
8 3.014 was replaced to 140 2.726 in same group
71 6.232 was replaced to 152 5.867 in same group
7 -0.089 was replaced to 177 -0.258 in same group
54 -4.336 was replaced to 179 -4.395 in same group
34 4.402 was replaced to 211 3.893 in same group
71 5.867 was replaced to 215 5.709 in same group
13 3.503 was replaced to 227 2.837 in same group
79 0.948 was replaced to 236 0.829 in same group
Bank No.: Energy
Bank 1: -6.232
Bank 2: -8.143
Bank 3: -11.490
Bank 4: -15.768
Bank 5: -17.438
Bank 6: -12.428
Bank 7: -16.942
Bank 8: -15.361
Bank 9: -6.440
Bank 10: -6.531
Bank 11: -10.754
Bank 12: -14.173
Bank 13: -14.917
Bank 14: -17.096
Bank 15: -3.817
Bank 16: 0.329
Bank 17: -12.520
Bank 18: -12.484
Bank 19: -18.843
Bank 20: -1.594
Bank 21: -9.337
Bank 22: 8.983
Bank 23: -16.139
Bank 24: -6.850
Bank 25: 2.476
Bank 26: -18.109
Bank 27: 1.413
Bank 28: -13.952
Bank 29: -15.404
Bank 30: -1.944
Bank 31: -12.930
Bank 32: -0.261
Bank 33: -17.383
Bank 34: 3.828
Bank 35: -16.676
Bank 36: -17.961
Bank 37: -1.174
Bank 38: -14.482
Bank 39: -10.748
Bank 40: -15.799
Bank 41: -15.273
Bank 42: -10.572
Bank 43: -7.233
Bank 44: -7.125
Bank 45: -15.998
Bank 46: -2.847
Bank 47: -11.080
Bank 48: -18.291
Bank 49: -3.911
Bank 50: 9.560
Bank 51: -7.725
Bank 52: -12.343
Bank 53: -8.530
Bank 54: -18.588
Bank 55: -10.692
Bank 56: -13.383
Bank 57: -17.716
Bank 58: -12.285
Bank 59: -10.040
Bank 60: -9.144
Bank 61: -16.307
Bank 62: -9.283
Bank 63: -14.577
Bank 64: -17.312
Bank 65: -19.571
Bank 66: -12.305
Bank 67: -12.252
Bank 68: -9.073
Bank 69: -12.832
Bank 70: -15.190
Bank 71: -13.358
Bank 72: -19.086
Bank 73: -18.355
Bank 74: -11.687
Bank 75: -9.666
Bank 76: -8.720
Bank 77: -16.726
Bank 78: -4.390
Bank 79: -17.907
Bank 80: -13.944
Bank 81: -10.018
Bank 82: -11.243
Bank 83: -13.821
Bank 84: -17.026
Bank 85: -5.422
Bank 86: -12.399
Bank 87: -11.549
Bank 88: -16.065
Bank 89: -17.945
Bank 90: -9.604
Bank 91: -19.070
Bank 92: -15.274
Bank 93: 5.350
Bank 94: -12.657
Bank 95: -18.945
Bank 96: -19.477
Bank 97: -18.377
Bank 98: -12.528
Bank 99: -10.422
Bank 100: -11.007
------------------------------------------------------------
CSA Run Number / Interation / SEED CYCLE = 1 / 1 / 62
D_ave / D_cut / D_min = 5.655 / 2.078 / 2.078
------------------------------------------------------------
Used time for CSA cycle 0D 0H 50M 41.7S
LCSA PLD: 1 0 15350 2.078 65 21 -19.571 -9.337 3765570 7 100 3041.749 sec
------------------------------------------------------------
Select Seeds....
Unused seeds : 7 / 100
------------------------------------------------------------
Make new conformation
perturbation rate : 0.000
calc_ml_energy.mol2 is working!
Time for inference: 41.36 (s)
21 7.046 was replaced to 57 6.659 in new group
36 -0.709 was replaced to 65 -1.710 in same group
18 6.199 was replaced to 114 3.113 in same group
98 6.479 was replaced to 117 6.096 in same group
13 2.837 was replaced to 130 2.667 in same group
79 0.829 was replaced to 139 0.142 in same group
84 3.818 was replaced to 143 3.798 in same group
45 -1.150 was replaced to 204 -1.232 in same group
29 5.693 was replaced to 209 5.581 in same group
Bank No.: Energy
Bank 1: -6.232
Bank 2: -8.143
Bank 3: -11.490
Bank 4: -15.768
Bank 5: -17.438
Bank 6: -12.428
Bank 7: -16.942
Bank 8: -15.361
Bank 9: -6.440
Bank 10: -6.531
Bank 11: -10.754
Bank 12: -14.173
Bank 13: -14.767
Bank 14: -17.096
Bank 15: -3.817
Bank 16: 0.329
Bank 17: -12.520
Bank 18: -16.021
Bank 19: -18.843
Bank 20: -1.594
Bank 21: 80.859
Bank 22: 8.983
Bank 23: -16.139
Bank 24: -6.850
Bank 25: 2.476
Bank 26: -18.109
Bank 27: 1.413
Bank 28: -13.952
Bank 29: -13.955
Bank 30: -1.944
Bank 31: -12.930
Bank 32: -0.261
Bank 33: -17.383
Bank 34: 3.828
Bank 35: -16.676
Bank 36: -16.235
Bank 37: -1.174
Bank 38: -14.482
Bank 39: -10.748
Bank 40: -15.799
Bank 41: -15.273
Bank 42: -10.572
Bank 43: -7.233
Bank 44: -7.125
Bank 45: -16.241
Bank 46: -2.847
Bank 47: -11.080
Bank 48: -18.291
Bank 49: -3.911
Bank 50: 9.560
Bank 51: -7.725
Bank 52: -12.343
Bank 53: -8.530
Bank 54: -18.588
Bank 55: -10.692
Bank 56: -13.383
Bank 57: -17.716
Bank 58: -12.285
Bank 59: -10.040
Bank 60: -9.144
Bank 61: -16.307
Bank 62: -9.283
Bank 63: -14.577
Bank 64: -17.312
Bank 65: -19.571
Bank 66: -12.305
Bank 67: -12.252
Bank 68: -9.073
Bank 69: -12.832
Bank 70: -15.190
Bank 71: -13.358
Bank 72: -19.086
Bank 73: -18.355
Bank 74: -11.687
Bank 75: -9.666
Bank 76: -8.720
Bank 77: -16.726
Bank 78: -4.390
Bank 79: -14.724
Bank 80: -13.944
Bank 81: -10.018
Bank 82: -11.243
Bank 83: -13.821
Bank 84: -13.980
Bank 85: -5.422
Bank 86: -12.399
Bank 87: -11.549
Bank 88: -16.065
Bank 89: -17.945
Bank 90: -9.604
Bank 91: -19.070
Bank 92: -15.274
Bank 93: 5.350
Bank 94: -12.657
Bank 95: -18.945
Bank 96: -19.477
Bank 97: -18.377
Bank 98: -9.926
Bank 99: -10.422
Bank 100: -11.007
------------------------------------------------------------
CSA Run Number / Interation / SEED CYCLE = 1 / 1 / 63
D_ave / D_cut / D_min = 5.628 / 2.078 / 2.078
------------------------------------------------------------
Used time for CSA cycle 0D 0H 51M 32.6S
LCSA PLD: 1 0 15600 2.078 65 99 -19.571 -10.422 3824167 9 100 3092.565 sec
------------------------------------------------------------
Select Seeds....
Unused seeds : 9 / 100
------------------------------------------------------------
Make new conformation
perturbation rate : 0.000
calc_ml_energy.mol2 is working!
Time for inference: 41.88 (s)
19 -0.474 was replaced to 6 -2.211 in same group
13 2.667 was replaced to 52 -0.204 in same group
18 3.113 was replaced to 54 2.684 in same group
38 6.083 was replaced to 153 5.800 in same group
61 5.027 was replaced to 197 4.236 in same group
33 1.463 was replaced to 205 1.127 in same group
1 5.223 was replaced to 213 4.952 in same group
96 -2.576 was replaced to 240 -2.656 in same group
Bank No.: Energy
Bank 1: -12.177
Bank 2: -8.143
Bank 3: -11.490
Bank 4: -15.768
Bank 5: -17.438
Bank 6: -12.428
Bank 7: -16.942
Bank 8: -15.361
Bank 9: -6.440
Bank 10: -6.531
Bank 11: -10.754
Bank 12: -14.173
Bank 13: -16.164
Bank 14: -17.096
Bank 15: -3.817
Bank 16: 0.329
Bank 17: -12.520
Bank 18: -8.539
Bank 19: -18.037
Bank 20: -1.594
Bank 21: 80.859
Bank 22: 8.983
Bank 23: -16.139
Bank 24: -6.850
Bank 25: 2.476
Bank 26: -18.109
Bank 27: 1.413
Bank 28: -13.952
Bank 29: -13.955
Bank 30: -1.944
Bank 31: -12.930
Bank 32: -0.261
Bank 33: -17.803
Bank 34: 3.828
Bank 35: -16.676
Bank 36: -16.235
Bank 37: -1.174
Bank 38: -15.511
Bank 39: -10.748
Bank 40: -15.799
Bank 41: -15.273
Bank 42: -10.572
Bank 43: -7.233
Bank 44: -7.125
Bank 45: -16.241
Bank 46: -2.847
Bank 47: -11.080
Bank 48: -18.291
Bank 49: -3.911
Bank 50: 9.560
Bank 51: -7.725
Bank 52: -12.343
Bank 53: -8.530
Bank 54: -18.588
Bank 55: -10.692
Bank 56: -13.383
Bank 57: -17.716
Bank 58: -12.285
Bank 59: -10.040
Bank 60: -9.144
Bank 61: -16.342
Bank 62: -9.283
Bank 63: -14.577
Bank 64: -17.312
Bank 65: -19.571
Bank 66: -12.305
Bank 67: -12.252
Bank 68: -9.073
Bank 69: -12.832
Bank 70: -15.190
Bank 71: -13.358
Bank 72: -19.086
Bank 73: -18.355
Bank 74: -11.687
Bank 75: -9.666
Bank 76: -8.720
Bank 77: -16.726
Bank 78: -4.390
Bank 79: -14.724
Bank 80: -13.944
Bank 81: -10.018
Bank 82: -11.243
Bank 83: -13.821
Bank 84: -13.980
Bank 85: -5.422
Bank 86: -12.399
Bank 87: -11.549
Bank 88: -16.065
Bank 89: -17.945
Bank 90: -9.604
Bank 91: -19.070
Bank 92: -15.274
Bank 93: 5.350
Bank 94: -12.657
Bank 95: -18.945
Bank 96: -19.866
Bank 97: -18.377
Bank 98: -9.926
Bank 99: -10.422
Bank 100: -11.007
------------------------------------------------------------
CSA Run Number / Interation / SEED CYCLE = 1 / 1 / 64
D_ave / D_cut / D_min = 5.612 / 2.078 / 2.078
------------------------------------------------------------
Used time for CSA cycle 0D 0H 52M 24.3S
LCSA PLD: 1 0 15850 2.078 65 99 -19.571 -10.422 3885169 8 100 3144.316 sec
------------------------------------------------------------
Select Seeds....
Unused seeds : 8 / 100
------------------------------------------------------------
Make new conformation
perturbation rate : 0.000
calc_ml_energy.mol2 is working!
Time for inference: 41.38 (s)
21 6.659 was replaced to 5 5.333 in same group
19 -2.211 was replaced to 8 -2.287 in same group
1 4.952 was replaced to 53 4.236 in same group
13 -0.204 was replaced to 54 -0.600 in same group
99 7.042 was replaced to 62 6.697 in new group
18 2.684 was replaced to 183 1.190 in same group
Bank No.: Energy
Bank 1: -12.537
Bank 2: -8.143
Bank 3: -11.490
Bank 4: -15.768
Bank 5: -17.438
Bank 6: -12.428
Bank 7: -16.942
Bank 8: -15.361
Bank 9: -6.440
Bank 10: -6.531
Bank 11: -10.754
Bank 12: -14.173
Bank 13: -16.961
Bank 14: -17.096
Bank 15: -3.817
Bank 16: 0.329
Bank 17: -12.520
Bank 18: -17.627
Bank 19: -17.639
Bank 20: -1.594
Bank 21: -12.607
Bank 22: 8.983
Bank 23: -16.139
Bank 24: -6.850
Bank 25: 2.476
Bank 26: -18.109
Bank 27: 1.413
Bank 28: -13.952
Bank 29: -13.955
Bank 30: -1.944
Bank 31: -12.930
Bank 32: -0.261
Bank 33: -17.803
Bank 34: 3.828
Bank 35: -16.676
Bank 36: -16.235
Bank 37: -1.174
Bank 38: -15.511
Bank 39: -10.748
Bank 40: -15.799
Bank 41: -15.273
Bank 42: -10.572
Bank 43: -7.233
Bank 44: -7.125
Bank 45: -16.241
Bank 46: -2.847
Bank 47: -11.080
Bank 48: -18.291
Bank 49: -3.911
Bank 50: 9.560
Bank 51: -7.725
Bank 52: -12.343
Bank 53: -8.530
Bank 54: -18.588
Bank 55: -10.692
Bank 56: -13.383
Bank 57: -17.716
Bank 58: -12.285
Bank 59: -10.040
Bank 60: -9.144
Bank 61: -16.342
Bank 62: -9.283
Bank 63: -14.577
Bank 64: -17.312
Bank 65: -19.571
Bank 66: -12.305
Bank 67: -12.252
Bank 68: -9.073
Bank 69: -12.832
Bank 70: -15.190
Bank 71: -13.358
Bank 72: -19.086
Bank 73: -18.355
Bank 74: -11.687
Bank 75: -9.666
Bank 76: -8.720
Bank 77: -16.726
Bank 78: -4.390
Bank 79: -14.724
Bank 80: -13.944
Bank 81: -10.018
Bank 82: -11.243
Bank 83: -13.821
Bank 84: -13.980
Bank 85: -5.422
Bank 86: -12.399
Bank 87: -11.549
Bank 88: -16.065
Bank 89: -17.945
Bank 90: -9.604
Bank 91: -19.070
Bank 92: -15.274
Bank 93: 5.350
Bank 94: -12.657
Bank 95: -18.945
Bank 96: -19.866
Bank 97: -18.377
Bank 98: -9.926
Bank 99: 1.017
Bank 100: -11.007
------------------------------------------------------------
CSA Run Number / Interation / SEED CYCLE = 1 / 1 / 65
D_ave / D_cut / D_min = 5.587 / 2.078 / 2.078
------------------------------------------------------------
Used time for CSA cycle 0D 0H 53M 14.8S
LCSA PLD: 1 0 16100 2.078 65 52 -19.571 -12.343 3943195 6 100 3194.816 sec
------------------------------------------------------------
Select Seeds....
Unused seeds : 6 / 100
------------------------------------------------------------
Make new conformation
perturbation rate : 0.000
calc_ml_energy.mol2 is working!
Time for inference: 41.83 (s)
99 6.697 was replaced to 136 4.483 in same group
1 4.236 was replaced to 176 3.297 in same group
48 1.014 was replaced to 212 0.287 in same group
71 5.709 was replaced to 238 5.262 in same group
26 1.137 was replaced to 242 0.950 in same group
Bank No.: Energy
Bank 1: -13.960
Bank 2: -8.143
Bank 3: -11.490
Bank 4: -15.768
Bank 5: -17.438
Bank 6: -12.428
Bank 7: -16.942
Bank 8: -15.361
Bank 9: -6.440
Bank 10: -6.531
Bank 11: -10.754
Bank 12: -14.173
Bank 13: -16.961
Bank 14: -17.096
Bank 15: -3.817
Bank 16: 0.329
Bank 17: -12.520
Bank 18: -17.627
Bank 19: -17.639
Bank 20: -1.594
Bank 21: -12.607
Bank 22: 8.983
Bank 23: -16.139
Bank 24: -6.850
Bank 25: 2.476
Bank 26: -17.646
Bank 27: 1.413
Bank 28: -13.952
Bank 29: -13.955
Bank 30: -1.944
Bank 31: -12.930
Bank 32: -0.261
Bank 33: -17.803
Bank 34: 3.828
Bank 35: -16.676
Bank 36: -16.235
Bank 37: -1.174
Bank 38: -15.511
Bank 39: -10.748
Bank 40: -15.799
Bank 41: -15.273
Bank 42: -10.572
Bank 43: -7.233
Bank 44: -7.125
Bank 45: -16.241
Bank 46: -2.847
Bank 47: -11.080
Bank 48: -19.933
Bank 49: -3.911
Bank 50: 9.560
Bank 51: -7.725
Bank 52: -12.343
Bank 53: -8.530
Bank 54: -18.588
Bank 55: -10.692
Bank 56: -13.383
Bank 57: -17.716
Bank 58: -12.285
Bank 59: -10.040
Bank 60: -9.144
Bank 61: -16.342
Bank 62: -9.283
Bank 63: -14.577
Bank 64: -17.312
Bank 65: -19.571
Bank 66: -12.305
Bank 67: -12.252
Bank 68: -9.073
Bank 69: -12.832
Bank 70: -15.190
Bank 71: -14.920
Bank 72: -19.086
Bank 73: -18.355
Bank 74: -11.687
Bank 75: -9.666
Bank 76: -8.720
Bank 77: -16.726
Bank 78: -4.390
Bank 79: -14.724
Bank 80: -13.944
Bank 81: -10.018
Bank 82: -11.243
Bank 83: -13.821
Bank 84: -13.980
Bank 85: -5.422
Bank 86: -12.399
Bank 87: -11.549
Bank 88: -16.065
Bank 89: -17.945
Bank 90: -9.604
Bank 91: -19.070
Bank 92: -15.274
Bank 93: 5.350
Bank 94: -12.657
Bank 95: -18.945
Bank 96: -19.866
Bank 97: -18.377
Bank 98: -9.926
Bank 99: -1.090
Bank 100: -11.007
------------------------------------------------------------
CSA Run Number / Interation / SEED CYCLE = 1 / 1 / 66
D_ave / D_cut / D_min = 5.585 / 2.078 / 2.078
------------------------------------------------------------
Used time for CSA cycle 0D 0H 54M 5.2S
LCSA PLD: 1 0 16350 2.078 65 52 -19.571 -12.343 4001246 5 100 3245.166 sec
------------------------------------------------------------
Select Seeds....
Unused seeds : 5 / 100
------------------------------------------------------------
Make new conformation
perturbation rate : 0.000
calc_ml_energy.mol2 is working!
Time for inference: 42.12 (s)
99 4.483 was replaced to 65 2.717 in same group
12 3.385 was replaced to 92 3.240 in same group
52 6.962 was replaced to 103 6.856 in new group
71 5.262 was replaced to 132 4.998 in same group
61 4.236 was replaced to 197 3.972 in same group
29 5.581 was replaced to 210 5.516 in same group
36 -1.710 was replaced to 213 -3.583 in same group
21 5.333 was replaced to 239 2.966 in same group
Bank No.: Energy
Bank 1: -13.960
Bank 2: -8.143
Bank 3: -11.490
Bank 4: -15.768
Bank 5: -17.438
Bank 6: -12.428
Bank 7: -16.942
Bank 8: -15.361
Bank 9: -6.440
Bank 10: -6.531
Bank 11: -10.754
Bank 12: -14.387
Bank 13: -16.961
Bank 14: -17.096
Bank 15: -3.817
Bank 16: 0.329
Bank 17: -12.520
Bank 18: -17.627
Bank 19: -17.639
Bank 20: -1.594
Bank 21: -11.537
Bank 22: 8.983
Bank 23: -16.139
Bank 24: -6.850
Bank 25: 2.476
Bank 26: -17.646
Bank 27: 1.413
Bank 28: -13.952
Bank 29: -15.568
Bank 30: -1.944
Bank 31: -12.930
Bank 32: -0.261
Bank 33: -17.803
Bank 34: 3.828
Bank 35: -16.676
Bank 36: -18.783
Bank 37: -1.174
Bank 38: -15.511
Bank 39: -10.748
Bank 40: -15.799
Bank 41: -15.273
Bank 42: -10.572
Bank 43: -7.233
Bank 44: -7.125
Bank 45: -16.241
Bank 46: -2.847
Bank 47: -11.080
Bank 48: -19.933
Bank 49: -3.911
Bank 50: 9.560
Bank 51: -7.725
Bank 52: -9.401
Bank 53: -8.530
Bank 54: -18.588
Bank 55: -10.692
Bank 56: -13.383
Bank 57: -17.716
Bank 58: -12.285
Bank 59: -10.040
Bank 60: -9.144
Bank 61: -9.285
Bank 62: -9.283
Bank 63: -14.577
Bank 64: -17.312
Bank 65: -19.571
Bank 66: -12.305
Bank 67: -12.252
Bank 68: -9.073
Bank 69: -12.832
Bank 70: -15.190
Bank 71: -11.043
Bank 72: -19.086
Bank 73: -18.355
Bank 74: -11.687
Bank 75: -9.666
Bank 76: -8.720
Bank 77: -16.726
Bank 78: -4.390
Bank 79: -14.724
Bank 80: -13.944
Bank 81: -10.018
Bank 82: -11.243
Bank 83: -13.821
Bank 84: -13.980
Bank 85: -5.422
Bank 86: -12.399
Bank 87: -11.549
Bank 88: -16.065
Bank 89: -17.945
Bank 90: -9.604
Bank 91: -19.070
Bank 92: -15.274
Bank 93: 5.350
Bank 94: -12.657
Bank 95: -18.945
Bank 96: -19.866
Bank 97: -18.377
Bank 98: -9.926
Bank 99: -12.246
Bank 100: -11.007
------------------------------------------------------------
CSA Run Number / Interation / SEED CYCLE = 1 / 1 / 67
D_ave / D_cut / D_min = 5.551 / 2.078 / 2.078
------------------------------------------------------------
Used time for CSA cycle 0D 0H 54M 55.5S
LCSA PLD: 1 0 16600 2.078 65 66 -19.571 -12.305 4063511 8 100 3295.452 sec
------------------------------------------------------------
Select Seeds....
Unused seeds : 8 / 100
------------------------------------------------------------
Make new conformation
perturbation rate : 0.000
calc_ml_energy.mol2 is working!
Time for inference: 43.14 (s)
99 2.717 was replaced to 74 1.994 in same group
52 6.856 was replaced to 135 3.638 in same group
36 -3.583 was replaced to 187 -4.004 in same group
52 3.638 was replaced to 188 1.446 in same group
71 4.998 was replaced to 195 4.613 in same group
38 5.800 was replaced to 210 5.565 in same group
Bank No.: Energy
Bank 1: -13.960
Bank 2: -8.143
Bank 3: -11.490
Bank 4: -15.768
Bank 5: -17.438
Bank 6: -12.428
Bank 7: -16.942
Bank 8: -15.361
Bank 9: -6.440
Bank 10: -6.531
Bank 11: -10.754
Bank 12: -14.387
Bank 13: -16.961
Bank 14: -17.096
Bank 15: -3.817
Bank 16: 0.329
Bank 17: -12.520
Bank 18: -17.627
Bank 19: -17.639
Bank 20: -1.594
Bank 21: -11.537
Bank 22: 8.983
Bank 23: -16.139
Bank 24: -6.850
Bank 25: 2.476
Bank 26: -17.646
Bank 27: 1.413
Bank 28: -13.952
Bank 29: -15.568
Bank 30: -1.944
Bank 31: -12.930
Bank 32: -0.261
Bank 33: -17.803
Bank 34: 3.828
Bank 35: -16.676
Bank 36: -18.292
Bank 37: -1.174
Bank 38: -14.644
Bank 39: -10.748
Bank 40: -15.799
Bank 41: -15.273
Bank 42: -10.572
Bank 43: -7.233
Bank 44: -7.125
Bank 45: -16.241
Bank 46: -2.847
Bank 47: -11.080
Bank 48: -19.933
Bank 49: -3.911
Bank 50: 9.560
Bank 51: -7.725
Bank 52: -19.317
Bank 53: -8.530
Bank 54: -18.588
Bank 55: -10.692
Bank 56: -13.383
Bank 57: -17.716
Bank 58: -12.285
Bank 59: -10.040
Bank 60: -9.144
Bank 61: -9.285
Bank 62: -9.283
Bank 63: -14.577
Bank 64: -17.312
Bank 65: -19.571
Bank 66: -12.305
Bank 67: -12.252
Bank 68: -9.073
Bank 69: -12.832
Bank 70: -15.190
Bank 71: -10.848
Bank 72: -19.086
Bank 73: -18.355
Bank 74: -11.687
Bank 75: -9.666
Bank 76: -8.720
Bank 77: -16.726
Bank 78: -4.390
Bank 79: -14.724
Bank 80: -13.944
Bank 81: -10.018
Bank 82: -11.243
Bank 83: -13.821
Bank 84: -13.980
Bank 85: -5.422
Bank 86: -12.399
Bank 87: -11.549
Bank 88: -16.065
Bank 89: -17.945
Bank 90: -9.604
Bank 91: -19.070
Bank 92: -15.274
Bank 93: 5.350
Bank 94: -12.657
Bank 95: -18.945
Bank 96: -19.866
Bank 97: -18.377
Bank 98: -9.926
Bank 99: -13.620
Bank 100: -11.007
------------------------------------------------------------
CSA Run Number / Interation / SEED CYCLE = 1 / 1 / 68
D_ave / D_cut / D_min = 5.542 / 2.078 / 2.078
------------------------------------------------------------
Used time for CSA cycle 0D 0H 55M 45.8S
LCSA PLD: 1 0 16850 2.078 65 66 -19.571 -12.305 4126065 5 100 3345.813 sec
------------------------------------------------------------
Select Seeds....
Unused seeds : 5 / 100
------------------------------------------------------------
Make new conformation
perturbation rate : 0.000
calc_ml_energy.mol2 is working!
Time for inference: 42.27 (s)
54 -4.395 was replaced to 85 -4.562 in same group
99 1.994 was replaced to 188 1.556 in same group
79 0.142 was replaced to 226 -0.334 in same group
Bank No.: Energy
Bank 1: -13.960
Bank 2: -8.143
Bank 3: -11.490
Bank 4: -15.768
Bank 5: -17.438
Bank 6: -12.428
Bank 7: -16.942
Bank 8: -15.361
Bank 9: -6.440
Bank 10: -6.531
Bank 11: -10.754
Bank 12: -14.387
Bank 13: -16.961
Bank 14: -17.096
Bank 15: -3.817
Bank 16: 0.329
Bank 17: -12.520
Bank 18: -17.627
Bank 19: -17.639
Bank 20: -1.594
Bank 21: -11.537
Bank 22: 8.983
Bank 23: -16.139
Bank 24: -6.850
Bank 25: 2.476
Bank 26: -17.646
Bank 27: 1.413
Bank 28: -13.952
Bank 29: -15.568
Bank 30: -1.944
Bank 31: -12.930
Bank 32: -0.261
Bank 33: -17.803
Bank 34: 3.828
Bank 35: -16.676
Bank 36: -18.292
Bank 37: -1.174
Bank 38: -14.644
Bank 39: -10.748
Bank 40: -15.799
Bank 41: -15.273
Bank 42: -10.572
Bank 43: -7.233
Bank 44: -7.125
Bank 45: -16.241
Bank 46: -2.847
Bank 47: -11.080
Bank 48: -19.933
Bank 49: -3.911
Bank 50: 9.560
Bank 51: -7.725
Bank 52: -19.317
Bank 53: -8.530
Bank 54: -17.616
Bank 55: -10.692
Bank 56: -13.383
Bank 57: -17.716
Bank 58: -12.285
Bank 59: -10.040
Bank 60: -9.144
Bank 61: -9.285
Bank 62: -9.283
Bank 63: -14.577
Bank 64: -17.312
Bank 65: -19.571
Bank 66: -12.305
Bank 67: -12.252
Bank 68: -9.073
Bank 69: -12.832
Bank 70: -15.190
Bank 71: -10.848
Bank 72: -19.086
Bank 73: -18.355
Bank 74: -11.687
Bank 75: -9.666
Bank 76: -8.720
Bank 77: -16.726
Bank 78: -4.390
Bank 79: -18.778
Bank 80: -13.944
Bank 81: -10.018
Bank 82: -11.243
Bank 83: -13.821
Bank 84: -13.980
Bank 85: -5.422
Bank 86: -12.399
Bank 87: -11.549
Bank 88: -16.065
Bank 89: -17.945
Bank 90: -9.604
Bank 91: -19.070
Bank 92: -15.274
Bank 93: 5.350
Bank 94: -12.657
Bank 95: -18.945
Bank 96: -19.866
Bank 97: -18.377
Bank 98: -9.926
Bank 99: -15.975
Bank 100: -11.007
------------------------------------------------------------
CSA Run Number / Interation / SEED CYCLE = 1 / 1 / 69
D_ave / D_cut / D_min = 5.541 / 2.078 / 2.078
------------------------------------------------------------
Used time for CSA cycle 0D 0H 56M 35.3S
LCSA PLD: 1 0 17100 2.078 54 66 -17.616 -12.305 4189382 3 100 3395.251 sec
------------------------------------------------------------
Select Seeds....
Unused seeds : 3 / 100
------------------------------------------------------------
Make new conformation
perturbation rate : 0.000
calc_ml_energy.mol2 is working!
Time for inference: 42.61 (s)
54 -4.562 was replaced to 177 -4.731 in same group
92 -3.162 was replaced to 178 -3.788 in same group
79 -0.334 was replaced to 180 -0.664 in same group
26 0.950 was replaced to 215 0.680 in same group
Bank No.: Energy
Bank 1: -13.960
Bank 2: -8.143
Bank 3: -11.490
Bank 4: -15.768
Bank 5: -17.438
Bank 6: -12.428
Bank 7: -16.942
Bank 8: -15.361
Bank 9: -6.440
Bank 10: -6.531
Bank 11: -10.754
Bank 12: -14.387
Bank 13: -16.961
Bank 14: -17.096
Bank 15: -3.817
Bank 16: 0.329
Bank 17: -12.520
Bank 18: -17.627
Bank 19: -17.639
Bank 20: -1.594
Bank 21: -11.537
Bank 22: 8.983
Bank 23: -16.139
Bank 24: -6.850
Bank 25: 2.476
Bank 26: -18.131
Bank 27: 1.413
Bank 28: -13.952
Bank 29: -15.568
Bank 30: -1.944
Bank 31: -12.930
Bank 32: -0.261
Bank 33: -17.803
Bank 34: 3.828
Bank 35: -16.676
Bank 36: -18.292
Bank 37: -1.174
Bank 38: -14.644
Bank 39: -10.748
Bank 40: -15.799
Bank 41: -15.273
Bank 42: -10.572
Bank 43: -7.233
Bank 44: -7.125
Bank 45: -16.241
Bank 46: -2.847
Bank 47: -11.080
Bank 48: -19.933
Bank 49: -3.911
Bank 50: 9.560
Bank 51: -7.725
Bank 52: -19.317
Bank 53: -8.530
Bank 54: -18.000
Bank 55: -10.692
Bank 56: -13.383
Bank 57: -17.716
Bank 58: -12.285
Bank 59: -10.040
Bank 60: -9.144
Bank 61: -9.285
Bank 62: -9.283
Bank 63: -14.577
Bank 64: -17.312
Bank 65: -19.571
Bank 66: -12.305
Bank 67: -12.252
Bank 68: -9.073
Bank 69: -12.832
Bank 70: -15.190
Bank 71: -10.848
Bank 72: -19.086
Bank 73: -18.355
Bank 74: -11.687
Bank 75: -9.666
Bank 76: -8.720
Bank 77: -16.726
Bank 78: -4.390
Bank 79: -18.321
Bank 80: -13.944
Bank 81: -10.018
Bank 82: -11.243
Bank 83: -13.821
Bank 84: -13.980
Bank 85: -5.422
Bank 86: -12.399
Bank 87: -11.549
Bank 88: -16.065
Bank 89: -17.945
Bank 90: -9.604
Bank 91: -19.070
Bank 92: -19.047
Bank 93: 5.350
Bank 94: -12.657
Bank 95: -18.945
Bank 96: -19.866
Bank 97: -18.377
Bank 98: -9.926
Bank 99: -15.975
Bank 100: -11.007
------------------------------------------------------------
CSA Run Number / Interation / SEED CYCLE = 1 / 1 / 70
D_ave / D_cut / D_min = 5.543 / 2.078 / 2.078
------------------------------------------------------------
Used time for CSA cycle 0D 0H 57M 25.1S
LCSA PLD: 1 0 17350 2.078 54 66 -18.000 -12.305 4250788 4 100 3445.081 sec
------------------------------------------------------------
Select Seeds....
Unused seeds : 4 / 100
------------------------------------------------------------
Make new conformation
perturbation rate : 0.000
calc_ml_energy.mol2 is working!
Time for inference: 43.52 (s)
79 -0.664 was replaced to 59 -0.837 in same group
52 1.446 was replaced to 88 0.066 in same group
88 -2.936 was replaced to 114 -3.344 in same group
83 6.713 was replaced to 158 6.507 in same group
66 6.946 was replaced to 175 6.379 in same group
99 1.556 was replaced to 222 1.004 in same group
17 6.716 was replaced to 244 6.052 in same group
Bank No.: Energy
Bank 1: -13.960
Bank 2: -8.143
Bank 3: -11.490
Bank 4: -15.768
Bank 5: -17.438
Bank 6: -12.428
Bank 7: -16.942
Bank 8: -15.361
Bank 9: -6.440
Bank 10: -6.531
Bank 11: -10.754
Bank 12: -14.387
Bank 13: -16.961
Bank 14: -17.096
Bank 15: -3.817
Bank 16: 0.329
Bank 17: -12.729
Bank 18: -17.627
Bank 19: -17.639
Bank 20: -1.594
Bank 21: -11.537
Bank 22: 8.983
Bank 23: -16.139
Bank 24: -6.850
Bank 25: 2.476
Bank 26: -18.131
Bank 27: 1.413
Bank 28: -13.952
Bank 29: -15.568
Bank 30: -1.944
Bank 31: -12.930
Bank 32: -0.261
Bank 33: -17.803
Bank 34: 3.828
Bank 35: -16.676
Bank 36: -18.292
Bank 37: -1.174
Bank 38: -14.644
Bank 39: -10.748
Bank 40: -15.799
Bank 41: -15.273
Bank 42: -10.572
Bank 43: -7.233
Bank 44: -7.125
Bank 45: -16.241
Bank 46: -2.847
Bank 47: -11.080
Bank 48: -19.933
Bank 49: -3.911
Bank 50: 9.560
Bank 51: -7.725
Bank 52: -19.246
Bank 53: -8.530
Bank 54: -18.000
Bank 55: -10.692
Bank 56: -13.383
Bank 57: -17.716
Bank 58: -12.285
Bank 59: -10.040
Bank 60: -9.144
Bank 61: -9.285
Bank 62: -9.283
Bank 63: -14.577
Bank 64: -17.312
Bank 65: -19.571
Bank 66: -11.255
Bank 67: -12.252
Bank 68: -9.073
Bank 69: -12.832
Bank 70: -15.190
Bank 71: -10.848
Bank 72: -19.086
Bank 73: -18.355
Bank 74: -11.687
Bank 75: -9.666
Bank 76: -8.720
Bank 77: -16.726
Bank 78: -4.390
Bank 79: -19.631
Bank 80: -13.944
Bank 81: -10.018
Bank 82: -11.243
Bank 83: -14.129
Bank 84: -13.980
Bank 85: -5.422
Bank 86: -12.399
Bank 87: -11.549
Bank 88: -15.776
Bank 89: -17.945
Bank 90: -9.604
Bank 91: -19.070
Bank 92: -19.047
Bank 93: 5.350
Bank 94: -12.657
Bank 95: -18.945
Bank 96: -19.866
Bank 97: -18.377
Bank 98: -9.926
Bank 99: -17.895
Bank 100: -11.007
------------------------------------------------------------
CSA Run Number / Interation / SEED CYCLE = 1 / 1 / 71
D_ave / D_cut / D_min = 5.547 / 2.078 / 2.078
------------------------------------------------------------
Used time for CSA cycle 0D 0H 58M 16.0S
LCSA PLD: 1 0 17600 2.078 54 2 -18.000 -8.143 4321425 7 100 3495.983 sec
------------------------------------------------------------
Select Seeds....
Unused seeds : 7 / 100
------------------------------------------------------------
Make new conformation
perturbation rate : 0.000
calc_ml_energy.mol2 is working!
Time for inference: 41.6 (s)
52 0.066 was replaced to 54 -0.370 in same group
54 -4.731 was replaced to 68 -5.666 in same group
64 1.149 was replaced to 101 0.578 in same group
45 -1.232 was replaced to 156 -1.280 in same group
52 -0.370 was replaced to 179 -0.451 in same group
79 -0.837 was replaced to 185 -1.358 in same group
Bank No.: Energy
Bank 1: -13.960
Bank 2: -8.143
Bank 3: -11.490
Bank 4: -15.768
Bank 5: -17.438
Bank 6: -12.428
Bank 7: -16.942
Bank 8: -15.361
Bank 9: -6.440
Bank 10: -6.531
Bank 11: -10.754
Bank 12: -14.387
Bank 13: -16.961
Bank 14: -17.096
Bank 15: -3.817
Bank 16: 0.329
Bank 17: -12.729
Bank 18: -17.627
Bank 19: -17.639
Bank 20: -1.594
Bank 21: -11.537
Bank 22: 8.983
Bank 23: -16.139
Bank 24: -6.850
Bank 25: 2.476
Bank 26: -18.131
Bank 27: 1.413
Bank 28: -13.952
Bank 29: -15.568
Bank 30: -1.944
Bank 31: -12.930
Bank 32: -0.261
Bank 33: -17.803
Bank 34: 3.828
Bank 35: -16.676
Bank 36: -18.292
Bank 37: -1.174
Bank 38: -14.644
Bank 39: -10.748
Bank 40: -15.799
Bank 41: -15.273
Bank 42: -10.572
Bank 43: -7.233
Bank 44: -7.125
Bank 45: -15.396
Bank 46: -2.847
Bank 47: -11.080
Bank 48: -19.933
Bank 49: -3.911
Bank 50: 9.560
Bank 51: -7.725
Bank 52: -19.388
Bank 53: -8.530
Bank 54: -18.657
Bank 55: -10.692
Bank 56: -13.383
Bank 57: -17.716
Bank 58: -12.285
Bank 59: -10.040
Bank 60: -9.144
Bank 61: -9.285
Bank 62: -9.283
Bank 63: -14.577
Bank 64: -9.407
Bank 65: -19.571
Bank 66: -11.255
Bank 67: -12.252
Bank 68: -9.073
Bank 69: -12.832
Bank 70: -15.190
Bank 71: -10.848
Bank 72: -19.086
Bank 73: -18.355
Bank 74: -11.687
Bank 75: -9.666
Bank 76: -8.720
Bank 77: -16.726
Bank 78: -4.390
Bank 79: -18.341
Bank 80: -13.944
Bank 81: -10.018
Bank 82: -11.243
Bank 83: -14.129
Bank 84: -13.980
Bank 85: -5.422
Bank 86: -12.399
Bank 87: -11.549
Bank 88: -15.776
Bank 89: -17.945
Bank 90: -9.604
Bank 91: -19.070
Bank 92: -19.047
Bank 93: 5.350
Bank 94: -12.657
Bank 95: -18.945
Bank 96: -19.866
Bank 97: -18.377
Bank 98: -9.926
Bank 99: -17.895
Bank 100: -11.007
------------------------------------------------------------
CSA Run Number / Interation / SEED CYCLE = 1 / 1 / 72
D_ave / D_cut / D_min = 5.542 / 2.078 / 2.078
------------------------------------------------------------
Used time for CSA cycle 0D 0H 59M 5.1S
LCSA PLD: 1 0 17850 2.078 54 2 -18.657 -8.143 4377957 5 100 3545.086 sec
------------------------------------------------------------
Select Seeds....
Unused seeds : 5 / 100
------------------------------------------------------------
Make new conformation
perturbation rate : 0.000
calc_ml_energy.mol2 is working!
Time for inference: 41.53 (s)
2 6.934 was replaced to 6 3.848 in new group
92 -3.788 was replaced to 130 -5.314 in same group
49 6.857 was replaced to 191 5.559 in new group
31 6.085 was replaced to 228 5.949 in same group
13 -0.600 was replaced to 247 -1.023 in same group
81 -0.292 was replaced to 249 -0.909 in same group
Bank No.: Energy
Bank 1: -13.960
Bank 2: -1.921
Bank 3: -11.490
Bank 4: -15.768
Bank 5: -17.438
Bank 6: -12.428
Bank 7: -16.942
Bank 8: -15.361
Bank 9: -6.440
Bank 10: -6.531
Bank 11: -10.754
Bank 12: -14.387
Bank 13: -18.596
Bank 14: -17.096
Bank 15: -3.817
Bank 16: 0.329
Bank 17: -12.729
Bank 18: -17.627
Bank 19: -17.639
Bank 20: -1.594
Bank 21: -11.537
Bank 22: 8.983
Bank 23: -16.139
Bank 24: -6.850
Bank 25: 2.476
Bank 26: -18.131
Bank 27: 1.413
Bank 28: -13.952
Bank 29: -15.568
Bank 30: -1.944
Bank 31: -14.267
Bank 32: -0.261
Bank 33: -17.803
Bank 34: 3.828
Bank 35: -16.676
Bank 36: -18.292
Bank 37: -1.174
Bank 38: -14.644
Bank 39: -10.748
Bank 40: -15.799
Bank 41: -15.273
Bank 42: -10.572
Bank 43: -7.233
Bank 44: -7.125
Bank 45: -15.396
Bank 46: -2.847
Bank 47: -11.080
Bank 48: -19.933
Bank 49: 2.398
Bank 50: 9.560
Bank 51: -7.725
Bank 52: -19.388
Bank 53: -8.530
Bank 54: -18.657
Bank 55: -10.692
Bank 56: -13.383
Bank 57: -17.716
Bank 58: -12.285
Bank 59: -10.040
Bank 60: -9.144
Bank 61: -9.285
Bank 62: -9.283
Bank 63: -14.577
Bank 64: -9.407
Bank 65: -19.571
Bank 66: -11.255
Bank 67: -12.252
Bank 68: -9.073
Bank 69: -12.832
Bank 70: -15.190
Bank 71: -10.848
Bank 72: -19.086
Bank 73: -18.355
Bank 74: -11.687
Bank 75: -9.666
Bank 76: -8.720
Bank 77: -16.726
Bank 78: -4.390
Bank 79: -18.341
Bank 80: -13.944
Bank 81: -12.171
Bank 82: -11.243
Bank 83: -14.129
Bank 84: -13.980
Bank 85: -5.422
Bank 86: -12.399
Bank 87: -11.549
Bank 88: -15.776
Bank 89: -17.945
Bank 90: -9.604
Bank 91: -19.070
Bank 92: -17.892
Bank 93: 5.350
Bank 94: -12.657
Bank 95: -18.945
Bank 96: -19.866
Bank 97: -18.377
Bank 98: -9.926
Bank 99: -17.895
Bank 100: -11.007
------------------------------------------------------------
CSA Run Number / Interation / SEED CYCLE = 1 / 1 / 73
D_ave / D_cut / D_min = 5.526 / 2.078 / 2.078
------------------------------------------------------------
Used time for CSA cycle 0D 0H 59M 54.3S
LCSA PLD: 1 0 18100 2.078 54 23 -18.657 -16.139 4433681 6 100 3594.280 sec
------------------------------------------------------------
Select Seeds....
Unused seeds : 6 / 100
------------------------------------------------------------
Make new conformation
perturbation rate : 0.000
calc_ml_energy.mol2 is working!
Time for inference: 41.19 (s)
10 2.947 was replaced to 95 2.234 in same group
2 3.848 was replaced to 127 3.457 in same group
90 -0.907 was replaced to 189 -1.069 in same group
20 4.608 was replaced to 247 4.529 in same group
Bank No.: Energy
Bank 1: -13.960
Bank 2: -8.816
Bank 3: -11.490
Bank 4: -15.768
Bank 5: -17.438
Bank 6: -12.428
Bank 7: -16.942
Bank 8: -15.361
Bank 9: -6.440
Bank 10: -11.074
Bank 11: -10.754
Bank 12: -14.387
Bank 13: -18.596
Bank 14: -17.096
Bank 15: -3.817
Bank 16: 0.329
Bank 17: -12.729
Bank 18: -17.627
Bank 19: -17.639
Bank 20: -14.290
Bank 21: -11.537
Bank 22: 8.983
Bank 23: -16.139
Bank 24: -6.850
Bank 25: 2.476
Bank 26: -18.131
Bank 27: 1.413
Bank 28: -13.952
Bank 29: -15.568
Bank 30: -1.944
Bank 31: -14.267
Bank 32: -0.261
Bank 33: -17.803
Bank 34: 3.828
Bank 35: -16.676
Bank 36: -18.292
Bank 37: -1.174
Bank 38: -14.644
Bank 39: -10.748
Bank 40: -15.799
Bank 41: -15.273
Bank 42: -10.572
Bank 43: -7.233
Bank 44: -7.125
Bank 45: -15.396
Bank 46: -2.847
Bank 47: -11.080
Bank 48: -19.933
Bank 49: 2.398
Bank 50: 9.560
Bank 51: -7.725
Bank 52: -19.388
Bank 53: -8.530
Bank 54: -18.657
Bank 55: -10.692
Bank 56: -13.383
Bank 57: -17.716
Bank 58: -12.285
Bank 59: -10.040
Bank 60: -9.144
Bank 61: -9.285
Bank 62: -9.283
Bank 63: -14.577
Bank 64: -9.407
Bank 65: -19.571
Bank 66: -11.255
Bank 67: -12.252
Bank 68: -9.073
Bank 69: -12.832
Bank 70: -15.190
Bank 71: -10.848
Bank 72: -19.086
Bank 73: -18.355
Bank 74: -11.687
Bank 75: -9.666
Bank 76: -8.720
Bank 77: -16.726
Bank 78: -4.390
Bank 79: -18.341
Bank 80: -13.944
Bank 81: -12.171
Bank 82: -11.243
Bank 83: -14.129
Bank 84: -13.980
Bank 85: -5.422
Bank 86: -12.399
Bank 87: -11.549
Bank 88: -15.776
Bank 89: -17.945
Bank 90: -0.102
Bank 91: -19.070
Bank 92: -17.892
Bank 93: 5.350
Bank 94: -12.657
Bank 95: -18.945
Bank 96: -19.866
Bank 97: -18.377
Bank 98: -9.926
Bank 99: -17.895
Bank 100: -11.007
------------------------------------------------------------
CSA Run Number / Interation / SEED CYCLE = 1 / 1 / 74
D_ave / D_cut / D_min = 5.512 / 2.078 / 2.078
------------------------------------------------------------
Used time for CSA cycle 0D 0H 60M 43.7S
LCSA PLD: 1 0 18350 2.078 54 23 -18.657 -16.139 4493182 4 100 3643.704 sec
------------------------------------------------------------
Select Seeds....
Unused seeds : 4 / 100
------------------------------------------------------------
Make new conformation
perturbation rate : 0.000
calc_ml_energy.mol2 is working!
Time for inference: 41.53 (s)
66 6.379 was replaced to 52 5.138 in same group
60 5.550 was replaced to 92 5.210 in same group
20 4.529 was replaced to 183 3.410 in same group
90 -1.069 was replaced to 187 -2.132 in same group
61 3.972 was replaced to 233 3.375 in same group
Bank No.: Energy
Bank 1: -13.960
Bank 2: -8.816
Bank 3: -11.490
Bank 4: -15.768
Bank 5: -17.438
Bank 6: -12.428
Bank 7: -16.942
Bank 8: -15.361
Bank 9: -6.440
Bank 10: -11.074
Bank 11: -10.754
Bank 12: -14.387
Bank 13: -18.596
Bank 14: -17.096
Bank 15: -3.817
Bank 16: 0.329
Bank 17: -12.729
Bank 18: -17.627
Bank 19: -17.639
Bank 20: -12.582
Bank 21: -11.537
Bank 22: 8.983
Bank 23: -16.139
Bank 24: -6.850
Bank 25: 2.476
Bank 26: -18.131
Bank 27: 1.413
Bank 28: -13.952
Bank 29: -15.568
Bank 30: -1.944
Bank 31: -14.267
Bank 32: -0.261
Bank 33: -17.803
Bank 34: 3.828
Bank 35: -16.676
Bank 36: -18.292
Bank 37: -1.174
Bank 38: -14.644
Bank 39: -10.748
Bank 40: -15.799
Bank 41: -15.273
Bank 42: -10.572
Bank 43: -7.233
Bank 44: -7.125
Bank 45: -15.396
Bank 46: -2.847
Bank 47: -11.080
Bank 48: -19.933
Bank 49: 2.398
Bank 50: 9.560
Bank 51: -7.725
Bank 52: -19.388
Bank 53: -8.530
Bank 54: -18.657
Bank 55: -10.692
Bank 56: -13.383
Bank 57: -17.716
Bank 58: -12.285
Bank 59: -10.040
Bank 60: -5.368
Bank 61: -16.728
Bank 62: -9.283
Bank 63: -14.577
Bank 64: -9.407
Bank 65: -19.571
Bank 66: -10.232
Bank 67: -12.252
Bank 68: -9.073
Bank 69: -12.832
Bank 70: -15.190
Bank 71: -10.848
Bank 72: -19.086
Bank 73: -18.355
Bank 74: -11.687
Bank 75: -9.666
Bank 76: -8.720
Bank 77: -16.726
Bank 78: -4.390
Bank 79: -18.341
Bank 80: -13.944
Bank 81: -12.171
Bank 82: -11.243
Bank 83: -14.129
Bank 84: -13.980
Bank 85: -5.422
Bank 86: -12.399
Bank 87: -11.549
Bank 88: -15.776
Bank 89: -17.945
Bank 90: -8.666
Bank 91: -19.070
Bank 92: -17.892
Bank 93: 5.350
Bank 94: -12.657
Bank 95: -18.945
Bank 96: -19.866
Bank 97: -18.377
Bank 98: -9.926
Bank 99: -17.895
Bank 100: -11.007
------------------------------------------------------------
CSA Run Number / Interation / SEED CYCLE = 1 / 1 / 75
D_ave / D_cut / D_min = 5.491 / 2.078 / 2.078
------------------------------------------------------------
Used time for CSA cycle 0D 1H 1M 33.7S
LCSA PLD: 1 0 18600 2.078 54 23 -18.657 -16.139 4553582 5 100 3693.724 sec
------------------------------------------------------------
Select Seeds....
Unused seeds : 5 / 100
------------------------------------------------------------
Make new conformation
perturbation rate : 0.000
calc_ml_energy.mol2 is working!
Time for inference: 39.95 (s)
23 6.734 was replaced to 10 4.266 in new group
15 6.638 was replaced to 63 4.795 in new group
40 -0.291 was replaced to 143 -0.673 in same group
20 3.410 was replaced to 178 3.191 in same group
Bank No.: Energy
Bank 1: -13.960
Bank 2: -8.816
Bank 3: -11.490
Bank 4: -15.768
Bank 5: -17.438
Bank 6: -12.428
Bank 7: -16.942
Bank 8: -15.361
Bank 9: -6.440
Bank 10: -11.074
Bank 11: -10.754
Bank 12: -14.387
Bank 13: -18.596
Bank 14: -17.096
Bank 15: -6.416
Bank 16: 0.329
Bank 17: -12.729
Bank 18: -17.627
Bank 19: -17.639
Bank 20: -12.513
Bank 21: -11.537
Bank 22: 8.983
Bank 23: -3.206
Bank 24: -6.850
Bank 25: 2.476
Bank 26: -18.131
Bank 27: 1.413
Bank 28: -13.952
Bank 29: -15.568
Bank 30: -1.944
Bank 31: -14.267
Bank 32: -0.261
Bank 33: -17.803
Bank 34: 3.828
Bank 35: -16.676
Bank 36: -18.292
Bank 37: -1.174
Bank 38: -14.644
Bank 39: -10.748
Bank 40: -15.058
Bank 41: -15.273
Bank 42: -10.572
Bank 43: -7.233
Bank 44: -7.125
Bank 45: -15.396
Bank 46: -2.847
Bank 47: -11.080
Bank 48: -19.933
Bank 49: 2.398
Bank 50: 9.560
Bank 51: -7.725
Bank 52: -19.388
Bank 53: -8.530
Bank 54: -18.657
Bank 55: -10.692
Bank 56: -13.383
Bank 57: -17.716
Bank 58: -12.285
Bank 59: -10.040
Bank 60: -5.368
Bank 61: -16.728
Bank 62: -9.283
Bank 63: -14.577
Bank 64: -9.407
Bank 65: -19.571
Bank 66: -10.232
Bank 67: -12.252
Bank 68: -9.073
Bank 69: -12.832
Bank 70: -15.190
Bank 71: -10.848
Bank 72: -19.086
Bank 73: -18.355
Bank 74: -11.687
Bank 75: -9.666
Bank 76: -8.720
Bank 77: -16.726
Bank 78: -4.390
Bank 79: -18.341
Bank 80: -13.944
Bank 81: -12.171
Bank 82: -11.243
Bank 83: -14.129
Bank 84: -13.980
Bank 85: -5.422
Bank 86: -12.399
Bank 87: -11.549
Bank 88: -15.776
Bank 89: -17.945
Bank 90: -8.666
Bank 91: -19.070
Bank 92: -17.892
Bank 93: 5.350
Bank 94: -12.657
Bank 95: -18.945
Bank 96: -19.866
Bank 97: -18.377
Bank 98: -9.926
Bank 99: -17.895
Bank 100: -11.007
------------------------------------------------------------
CSA Run Number / Interation / SEED CYCLE = 1 / 1 / 76
D_ave / D_cut / D_min = 5.445 / 2.078 / 2.078
------------------------------------------------------------
Used time for CSA cycle 0D 1H 2M 23.0S
LCSA PLD: 1 0 18850 2.078 54 30 -18.657 -1.944 4612025 4 100 3743.000 sec
------------------------------------------------------------
Select Seeds....
Unused seeds : 4 / 100
------------------------------------------------------------
Make new conformation
perturbation rate : 0.000
calc_ml_energy.mol2 is working!
Time for inference: 40.39 (s)
70 0.654 was replaced to 170 0.357 in same group
30 6.607 was replaced to 196 6.461 in new group
7 -0.258 was replaced to 203 -0.881 in same group
4 -0.571 was replaced to 236 -0.758 in same group
99 1.004 was replaced to 247 0.755 in same group
Bank No.: Energy
Bank 1: -13.960
Bank 2: -8.816
Bank 3: -11.490
Bank 4: 23.381
Bank 5: -17.438
Bank 6: -12.428
Bank 7: -17.026
Bank 8: -15.361
Bank 9: -6.440
Bank 10: -11.074
Bank 11: -10.754
Bank 12: -14.387
Bank 13: -18.596
Bank 14: -17.096
Bank 15: -6.416
Bank 16: 0.329
Bank 17: -12.729
Bank 18: -17.627
Bank 19: -17.639
Bank 20: -12.513
Bank 21: -11.537
Bank 22: 8.983
Bank 23: -3.206
Bank 24: -6.850
Bank 25: 2.476
Bank 26: -18.131
Bank 27: 1.413
Bank 28: -13.952
Bank 29: -15.568
Bank 30: 104.432
Bank 31: -14.267
Bank 32: -0.261
Bank 33: -17.803
Bank 34: 3.828
Bank 35: -16.676
Bank 36: -18.292
Bank 37: -1.174
Bank 38: -14.644
Bank 39: -10.748
Bank 40: -15.058
Bank 41: -15.273
Bank 42: -10.572
Bank 43: -7.233
Bank 44: -7.125
Bank 45: -15.396
Bank 46: -2.847
Bank 47: -11.080
Bank 48: -19.933
Bank 49: 2.398
Bank 50: 9.560
Bank 51: -7.725
Bank 52: -19.388
Bank 53: -8.530
Bank 54: -18.657
Bank 55: -10.692
Bank 56: -13.383
Bank 57: -17.716
Bank 58: -12.285
Bank 59: -10.040
Bank 60: -5.368
Bank 61: -16.728
Bank 62: -9.283
Bank 63: -14.577
Bank 64: -9.407
Bank 65: -19.571
Bank 66: -10.232
Bank 67: -12.252
Bank 68: -9.073
Bank 69: -12.832
Bank 70: -16.505
Bank 71: -10.848
Bank 72: -19.086
Bank 73: -18.355
Bank 74: -11.687
Bank 75: -9.666
Bank 76: -8.720
Bank 77: -16.726
Bank 78: -4.390
Bank 79: -18.341
Bank 80: -13.944
Bank 81: -12.171
Bank 82: -11.243
Bank 83: -14.129
Bank 84: -13.980
Bank 85: -5.422
Bank 86: -12.399
Bank 87: -11.549
Bank 88: -15.776
Bank 89: -17.945
Bank 90: -8.666
Bank 91: -19.070
Bank 92: -17.892
Bank 93: 5.350
Bank 94: -12.657
Bank 95: -18.945
Bank 96: -19.866
Bank 97: -18.377
Bank 98: -9.926
Bank 99: -17.222
Bank 100: -11.007
------------------------------------------------------------
CSA Run Number / Interation / SEED CYCLE = 1 / 1 / 77
D_ave / D_cut / D_min = 5.404 / 2.078 / 2.078
------------------------------------------------------------
Used time for CSA cycle 0D 1H 3M 12.8S
LCSA PLD: 1 0 19100 2.078 54 22 -18.657 8.983 4675319 5 100 3792.799 sec
------------------------------------------------------------
Select Seeds....
Unused seeds : 5 / 100
------------------------------------------------------------
Make new conformation
perturbation rate : 0.000
calc_ml_energy.mol2 is working!
Time for inference: 40.8 (s)
8 2.726 was replaced to 19 2.601 in same group
22 6.602 was replaced to 39 6.363 in new group
52 -0.451 was replaced to 98 -2.183 in same group
99 0.755 was replaced to 135 0.718 in same group
31 5.949 was replaced to 225 5.743 in same group
Bank No.: Energy
Bank 1: -13.960
Bank 2: -8.816
Bank 3: -11.490
Bank 4: 23.381
Bank 5: -17.438
Bank 6: -12.428
Bank 7: -17.026
Bank 8: -15.261
Bank 9: -6.440
Bank 10: -11.074
Bank 11: -10.754
Bank 12: -14.387
Bank 13: -18.596
Bank 14: -17.096
Bank 15: -6.416
Bank 16: 0.329
Bank 17: -12.729
Bank 18: -17.627
Bank 19: -17.639
Bank 20: -12.513
Bank 21: -11.537
Bank 22: 65.013
Bank 23: -3.206
Bank 24: -6.850
Bank 25: 2.476
Bank 26: -18.131
Bank 27: 1.413
Bank 28: -13.952
Bank 29: -15.568
Bank 30: 104.432
Bank 31: -13.290
Bank 32: -0.261
Bank 33: -17.803
Bank 34: 3.828
Bank 35: -16.676
Bank 36: -18.292
Bank 37: -1.174
Bank 38: -14.644
Bank 39: -10.748
Bank 40: -15.058
Bank 41: -15.273
Bank 42: -10.572
Bank 43: -7.233
Bank 44: -7.125
Bank 45: -15.396
Bank 46: -2.847
Bank 47: -11.080
Bank 48: -19.933
Bank 49: 2.398
Bank 50: 9.560
Bank 51: -7.725
Bank 52: -19.312
Bank 53: -8.530
Bank 54: -18.657
Bank 55: -10.692
Bank 56: -13.383
Bank 57: -17.716
Bank 58: -12.285
Bank 59: -10.040
Bank 60: -5.368
Bank 61: -16.728
Bank 62: -9.283
Bank 63: -14.577
Bank 64: -9.407
Bank 65: -19.571
Bank 66: -10.232
Bank 67: -12.252
Bank 68: -9.073
Bank 69: -12.832
Bank 70: -16.505
Bank 71: -10.848
Bank 72: -19.086
Bank 73: -18.355
Bank 74: -11.687
Bank 75: -9.666
Bank 76: -8.720
Bank 77: -16.726
Bank 78: -4.390
Bank 79: -18.341
Bank 80: -13.944
Bank 81: -12.171
Bank 82: -11.243
Bank 83: -14.129
Bank 84: -13.980
Bank 85: -5.422
Bank 86: -12.399
Bank 87: -11.549
Bank 88: -15.776
Bank 89: -17.945
Bank 90: -8.666
Bank 91: -19.070
Bank 92: -17.892
Bank 93: 5.350
Bank 94: -12.657
Bank 95: -18.945
Bank 96: -19.866
Bank 97: -18.377
Bank 98: -9.926
Bank 99: -17.427
Bank 100: -11.007
------------------------------------------------------------
CSA Run Number / Interation / SEED CYCLE = 1 / 1 / 78
D_ave / D_cut / D_min = 5.365 / 2.078 / 2.078
------------------------------------------------------------
Used time for CSA cycle 0D 1H 4M 3.2S
LCSA PLD: 1 0 19350 2.078 54 37 -18.657 -1.174 4729339 5 100 3843.178 sec
------------------------------------------------------------
Select Seeds....
Unused seeds : 5 / 100
------------------------------------------------------------
Make new conformation
perturbation rate : 0.000
calc_ml_energy.mol2 is working!
Time for inference: 42.4 (s)
37 6.566 was replaced to 13 6.370 in new group
22 6.363 was replaced to 128 5.005 in same group
40 -0.673 was replaced to 222 -0.849 in same group
13 -1.023 was replaced to 224 -2.194 in same group
28 0.394 was replaced to 227 0.015 in same group
63 -0.087 was replaced to 232 -0.285 in same group
Bank No.: Energy
Bank 1: -13.960
Bank 2: -8.816
Bank 3: -11.490
Bank 4: 23.381
Bank 5: -17.438
Bank 6: -12.428
Bank 7: -17.026
Bank 8: -15.261
Bank 9: -6.440
Bank 10: -11.074
Bank 11: -10.754
Bank 12: -14.387
Bank 13: -19.119
Bank 14: -17.096
Bank 15: -6.416
Bank 16: 0.329
Bank 17: -12.729
Bank 18: -17.627
Bank 19: -17.639
Bank 20: -12.513
Bank 21: -11.537
Bank 22: 29.302
Bank 23: -3.206
Bank 24: -6.850
Bank 25: 2.476
Bank 26: -18.131
Bank 27: 1.413
Bank 28: -2.509
Bank 29: -15.568
Bank 30: 104.432
Bank 31: -13.290
Bank 32: -0.261
Bank 33: -17.803
Bank 34: 3.828
Bank 35: -16.676
Bank 36: -18.292
Bank 37: -4.745
Bank 38: -14.644
Bank 39: -10.748
Bank 40: -15.019
Bank 41: -15.273
Bank 42: -10.572
Bank 43: -7.233
Bank 44: -7.125
Bank 45: -15.396
Bank 46: -2.847
Bank 47: -11.080
Bank 48: -19.933
Bank 49: 2.398
Bank 50: 9.560
Bank 51: -7.725
Bank 52: -19.312
Bank 53: -8.530
Bank 54: -18.657
Bank 55: -10.692
Bank 56: -13.383
Bank 57: -17.716
Bank 58: -12.285
Bank 59: -10.040
Bank 60: -5.368
Bank 61: -16.728
Bank 62: -9.283
Bank 63: -16.858
Bank 64: -9.407
Bank 65: -19.571
Bank 66: -10.232
Bank 67: -12.252
Bank 68: -9.073
Bank 69: -12.832
Bank 70: -16.505
Bank 71: -10.848
Bank 72: -19.086
Bank 73: -18.355
Bank 74: -11.687
Bank 75: -9.666
Bank 76: -8.720
Bank 77: -16.726
Bank 78: -4.390
Bank 79: -18.341
Bank 80: -13.944
Bank 81: -12.171
Bank 82: -11.243
Bank 83: -14.129
Bank 84: -13.980
Bank 85: -5.422
Bank 86: -12.399
Bank 87: -11.549
Bank 88: -15.776
Bank 89: -17.945
Bank 90: -8.666
Bank 91: -19.070
Bank 92: -17.892
Bank 93: 5.350
Bank 94: -12.657
Bank 95: -18.945
Bank 96: -19.866
Bank 97: -18.377
Bank 98: -9.926
Bank 99: -17.427
Bank 100: -11.007
------------------------------------------------------------
CSA Run Number / Interation / SEED CYCLE = 1 / 1 / 79
D_ave / D_cut / D_min = 5.343 / 2.078 / 2.078
------------------------------------------------------------
Used time for CSA cycle 0D 1H 4M 54.8S
LCSA PLD: 1 0 19600 2.078 54 87 -18.657 -11.549 4791819 6 100 3894.769 sec
------------------------------------------------------------
Select Seeds....
Unused seeds : 6 / 100
------------------------------------------------------------
Make new conformation
perturbation rate : 0.000
calc_ml_energy.mol2 is working!
Time for inference: 42.66 (s)
48 0.287 was replaced to 97 -0.483 in same group
81 -0.909 was replaced to 157 -0.998 in same group
37 6.370 was replaced to 186 5.344 in same group
70 0.357 was replaced to 192 0.032 in same group
81 -0.998 was replaced to 221 -1.038 in same group
7 -0.881 was replaced to 223 -1.271 in same group
Bank No.: Energy
Bank 1: -13.960
Bank 2: -8.816
Bank 3: -11.490
Bank 4: 23.381
Bank 5: -17.438
Bank 6: -12.428
Bank 7: -17.246
Bank 8: -15.261
Bank 9: -6.440
Bank 10: -11.074
Bank 11: -10.754
Bank 12: -14.387
Bank 13: -19.119
Bank 14: -17.096
Bank 15: -6.416
Bank 16: 0.329
Bank 17: -12.729
Bank 18: -17.627
Bank 19: -17.639
Bank 20: -12.513
Bank 21: -11.537
Bank 22: 29.302
Bank 23: -3.206
Bank 24: -6.850
Bank 25: 2.476
Bank 26: -18.131
Bank 27: 1.413
Bank 28: -2.509
Bank 29: -15.568
Bank 30: 104.432
Bank 31: -13.290
Bank 32: -0.261
Bank 33: -17.803
Bank 34: 3.828
Bank 35: -16.676
Bank 36: -18.292
Bank 37: -6.621
Bank 38: -14.644
Bank 39: -10.748
Bank 40: -15.019
Bank 41: -15.273
Bank 42: -10.572
Bank 43: -7.233
Bank 44: -7.125
Bank 45: -15.396
Bank 46: -2.847
Bank 47: -11.080
Bank 48: -16.477
Bank 49: 2.398
Bank 50: 9.560
Bank 51: -7.725
Bank 52: -19.312
Bank 53: -8.530
Bank 54: -18.657
Bank 55: -10.692
Bank 56: -13.383
Bank 57: -17.716
Bank 58: -12.285
Bank 59: -10.040
Bank 60: -5.368
Bank 61: -16.728
Bank 62: -9.283
Bank 63: -16.858
Bank 64: -9.407
Bank 65: -19.571
Bank 66: -10.232
Bank 67: -12.252
Bank 68: -9.073
Bank 69: -12.832
Bank 70: -17.445
Bank 71: -10.848
Bank 72: -19.086
Bank 73: -18.355
Bank 74: -11.687
Bank 75: -9.666
Bank 76: -8.720
Bank 77: -16.726
Bank 78: -4.390
Bank 79: -18.341
Bank 80: -13.944
Bank 81: -16.000
Bank 82: -11.243
Bank 83: -14.129
Bank 84: -13.980
Bank 85: -5.422
Bank 86: -12.399
Bank 87: -11.549
Bank 88: -15.776
Bank 89: -17.945
Bank 90: -8.666
Bank 91: -19.070
Bank 92: -17.892
Bank 93: 5.350
Bank 94: -12.657
Bank 95: -18.945
Bank 96: -19.866
Bank 97: -18.377
Bank 98: -9.926
Bank 99: -17.427
Bank 100: -11.007
------------------------------------------------------------
CSA Run Number / Interation / SEED CYCLE = 1 / 1 / 80
D_ave / D_cut / D_min = 5.338 / 2.078 / 2.078
------------------------------------------------------------
Used time for CSA cycle 0D 1H 5M 44.9S
LCSA PLD: 1 0 19850 2.078 54 87 -18.657 -11.549 4852545 5 100 3944.908 sec
------------------------------------------------------------
Select Seeds....
Unused seeds : 5 / 100
------------------------------------------------------------
Make new conformation
perturbation rate : 0.000
calc_ml_energy.mol2 is working!
Time for inference: 43.52 (s)
50 4.718 was replaced to 119 3.168 in same group
Bank No.: Energy
Bank 1: -13.960
Bank 2: -8.816
Bank 3: -11.490
Bank 4: 23.381
Bank 5: -17.438
Bank 6: -12.428
Bank 7: -17.246
Bank 8: -15.261
Bank 9: -6.440
Bank 10: -11.074
Bank 11: -10.754
Bank 12: -14.387
Bank 13: -19.119
Bank 14: -17.096
Bank 15: -6.416
Bank 16: 0.329
Bank 17: -12.729
Bank 18: -17.627
Bank 19: -17.639
Bank 20: -12.513
Bank 21: -11.537
Bank 22: 29.302
Bank 23: -3.206
Bank 24: -6.850
Bank 25: 2.476
Bank 26: -18.131
Bank 27: 1.413
Bank 28: -2.509
Bank 29: -15.568
Bank 30: 104.432
Bank 31: -13.290
Bank 32: -0.261
Bank 33: -17.803
Bank 34: 3.828
Bank 35: -16.676
Bank 36: -18.292
Bank 37: -6.621
Bank 38: -14.644
Bank 39: -10.748
Bank 40: -15.019
Bank 41: -15.273
Bank 42: -10.572
Bank 43: -7.233
Bank 44: -7.125
Bank 45: -15.396
Bank 46: -2.847
Bank 47: -11.080
Bank 48: -16.477
Bank 49: 2.398
Bank 50: -9.384
Bank 51: -7.725
Bank 52: -19.312
Bank 53: -8.530
Bank 54: -18.657
Bank 55: -10.692
Bank 56: -13.383
Bank 57: -17.716
Bank 58: -12.285
Bank 59: -10.040
Bank 60: -5.368
Bank 61: -16.728
Bank 62: -9.283
Bank 63: -16.858
Bank 64: -9.407
Bank 65: -19.571
Bank 66: -10.232
Bank 67: -12.252
Bank 68: -9.073
Bank 69: -12.832
Bank 70: -17.445
Bank 71: -10.848
Bank 72: -19.086
Bank 73: -18.355
Bank 74: -11.687
Bank 75: -9.666
Bank 76: -8.720
Bank 77: -16.726
Bank 78: -4.390
Bank 79: -18.341
Bank 80: -13.944
Bank 81: -16.000
Bank 82: -11.243
Bank 83: -14.129
Bank 84: -13.980
Bank 85: -5.422
Bank 86: -12.399
Bank 87: -11.549
Bank 88: -15.776
Bank 89: -17.945
Bank 90: -8.666
Bank 91: -19.070
Bank 92: -17.892
Bank 93: 5.350
Bank 94: -12.657
Bank 95: -18.945
Bank 96: -19.866
Bank 97: -18.377
Bank 98: -9.926
Bank 99: -17.427
Bank 100: -11.007
------------------------------------------------------------
CSA Run Number / Interation / SEED CYCLE = 1 / 1 / 81
D_ave / D_cut / D_min = 5.329 / 2.078 / 2.078
------------------------------------------------------------
Used time for CSA cycle 0D 1H 6M 35.8S
LCSA PLD: 1 1 20100 2.078 54 87 -18.657 -11.549 4918205 100 100 3995.765 sec
------------------------------------------------------------
Select Seeds....
Unused seeds : 100 / 100
------------------------------------------------------------
Make new conformation
perturbation rate : 0.000
calc_ml_energy.mol2 is working!
Time for inference: 41.95 (s)
77 -3.084 was replaced to 62 -3.290 in same group
1 3.297 was replaced to 96 2.208 in same group
48 -0.483 was replaced to 148 -0.867 in same group
57 -1.399 was replaced to 187 -1.451 in same group
29 5.516 was replaced to 192 5.331 in same group
63 -0.285 was replaced to 232 -1.459 in same group
94 -0.361 was replaced to 249 -0.620 in same group
Bank No.: Energy
Bank 1: -14.693
Bank 2: -8.816
Bank 3: -11.490
Bank 4: 23.381
Bank 5: -17.438
Bank 6: -12.428
Bank 7: -17.246
Bank 8: -15.261
Bank 9: -6.440
Bank 10: -11.074
Bank 11: -10.754
Bank 12: -14.387
Bank 13: -19.119
Bank 14: -17.096
Bank 15: -6.416
Bank 16: 0.329
Bank 17: -12.729
Bank 18: -17.627
Bank 19: -17.639
Bank 20: -12.513
Bank 21: -11.537
Bank 22: 29.302
Bank 23: -3.206
Bank 24: -6.850
Bank 25: 2.476
Bank 26: -18.131
Bank 27: 1.413
Bank 28: -2.509
Bank 29: -16.706
Bank 30: 104.432
Bank 31: -13.290
Bank 32: -0.261
Bank 33: -17.803
Bank 34: 3.828
Bank 35: -16.676
Bank 36: -18.292
Bank 37: -6.621
Bank 38: -14.644
Bank 39: -10.748
Bank 40: -15.019
Bank 41: -15.273
Bank 42: -10.572
Bank 43: -7.233
Bank 44: -7.125
Bank 45: -15.396
Bank 46: -2.847
Bank 47: -11.080
Bank 48: -12.722
Bank 49: 2.398
Bank 50: -9.384
Bank 51: -7.725
Bank 52: -19.312
Bank 53: -8.530
Bank 54: -18.657
Bank 55: -10.692
Bank 56: -13.383
Bank 57: -18.551
Bank 58: -12.285
Bank 59: -10.040
Bank 60: -5.368
Bank 61: -16.728
Bank 62: -9.283
Bank 63: -15.991
Bank 64: -9.407
Bank 65: -19.571
Bank 66: -10.232
Bank 67: -12.252
Bank 68: -9.073
Bank 69: -12.832
Bank 70: -17.445
Bank 71: -10.848
Bank 72: -19.086
Bank 73: -18.355
Bank 74: -11.687
Bank 75: -9.666
Bank 76: -8.720
Bank 77: -16.693
Bank 78: -4.390
Bank 79: -18.341
Bank 80: -13.944
Bank 81: -16.000
Bank 82: -11.243
Bank 83: -14.129
Bank 84: -13.980
Bank 85: -5.422
Bank 86: -12.399
Bank 87: -11.549
Bank 88: -15.776
Bank 89: -17.945
Bank 90: -8.666
Bank 91: -19.070
Bank 92: -17.892
Bank 93: 5.350
Bank 94: -16.711
Bank 95: -18.945
Bank 96: -19.866
Bank 97: -18.377
Bank 98: -9.926
Bank 99: -17.427
Bank 100: -11.007
------------------------------------------------------------
CSA Run Number / Interation / SEED CYCLE = 1 / 1 / 82
D_ave / D_cut / D_min = 5.340 / 2.078 / 2.078
------------------------------------------------------------
Used time for CSA cycle 0D 1H 7M 25.0S
LCSA PLD: 1 1 20350 2.078 54 87 -18.657 -11.549 4984030 78 100 4045.010 sec
------------------------------------------------------------
Select Seeds....
Unused seeds : 78 / 100
------------------------------------------------------------
Make new conformation
perturbation rate : 0.000
calc_ml_energy.mol2 is working!
Time for inference: 44.21 (s)
6 4.785 was replaced to 72 4.561 in same group
87 6.539 was replaced to 104 5.836 in new group
93 6.011 was replaced to 119 5.176 in same group
1 2.208 was replaced to 168 1.789 in same group
65 -4.409 was replaced to 178 -5.023 in same group
84 3.798 was replaced to 191 3.476 in same group
35 0.507 was replaced to 205 -0.014 in same group
21 2.966 was replaced to 221 2.160 in same group
47 6.165 was replaced to 233 4.114 in same group
Bank No.: Energy
Bank 1: -15.626
Bank 2: -8.816
Bank 3: -11.490
Bank 4: 23.381
Bank 5: -17.438
Bank 6: -13.300
Bank 7: -17.246
Bank 8: -15.261
Bank 9: -6.440
Bank 10: -11.074
Bank 11: -10.754
Bank 12: -14.387
Bank 13: -19.119
Bank 14: -17.096
Bank 15: -6.416
Bank 16: 0.329
Bank 17: -12.729
Bank 18: -17.627
Bank 19: -17.639
Bank 20: -12.513
Bank 21: -13.274
Bank 22: 29.302
Bank 23: -3.206
Bank 24: -6.850
Bank 25: 2.476
Bank 26: -18.131
Bank 27: 1.413
Bank 28: -2.509
Bank 29: -16.706
Bank 30: 104.432
Bank 31: -13.290
Bank 32: -0.261
Bank 33: -17.803
Bank 34: 3.828
Bank 35: -18.140
Bank 36: -18.292
Bank 37: -6.621
Bank 38: -14.644
Bank 39: -10.748
Bank 40: -15.019
Bank 41: -15.273
Bank 42: -10.572
Bank 43: -7.233
Bank 44: -7.125
Bank 45: -15.396
Bank 46: -2.847
Bank 47: -11.380
Bank 48: -12.722
Bank 49: 2.398
Bank 50: -9.384
Bank 51: -7.725
Bank 52: -19.312
Bank 53: -8.530
Bank 54: -18.657
Bank 55: -10.692
Bank 56: -13.383
Bank 57: -18.551
Bank 58: -12.285
Bank 59: -10.040
Bank 60: -5.368
Bank 61: -16.728
Bank 62: -9.283
Bank 63: -15.991
Bank 64: -9.407
Bank 65: -20.055
Bank 66: -10.232
Bank 67: -12.252
Bank 68: -9.073
Bank 69: -12.832
Bank 70: -17.445
Bank 71: -10.848
Bank 72: -19.086
Bank 73: -18.355
Bank 74: -11.687
Bank 75: -9.666
Bank 76: -8.720
Bank 77: -16.693
Bank 78: -4.390
Bank 79: -18.341
Bank 80: -13.944
Bank 81: -16.000
Bank 82: -11.243
Bank 83: -14.129
Bank 84: -15.318
Bank 85: -5.422
Bank 86: -12.399
Bank 87: -4.529
Bank 88: -15.776
Bank 89: -17.945
Bank 90: -8.666
Bank 91: -19.070
Bank 92: -17.892
Bank 93: -9.427
Bank 94: -16.711
Bank 95: -18.945
Bank 96: -19.866
Bank 97: -18.377
Bank 98: -9.926
Bank 99: -17.427
Bank 100: -11.007
------------------------------------------------------------
CSA Run Number / Interation / SEED CYCLE = 1 / 1 / 83
D_ave / D_cut / D_min = 5.286 / 2.078 / 2.078
------------------------------------------------------------
Used time for CSA cycle 0D 1H 8M 16.6S
LCSA PLD: 1 1 20600 2.078 54 83 -18.657 -14.129 5043209 61 100 4096.595 sec
------------------------------------------------------------
Select Seeds....
Unused seeds : 61 / 100
------------------------------------------------------------
Make new conformation
perturbation rate : 0.000
calc_ml_energy.mol2 is working!
Time for inference: 41.14 (s)
83 6.507 was replaced to 54 5.822 in new group
75 6.020 was replaced to 116 5.799 in same group
93 5.176 was replaced to 118 1.138 in same group
94 -0.620 was replaced to 119 -1.336 in same group
47 4.114 was replaced to 137 3.563 in same group
21 2.160 was replaced to 152 1.394 in same group
5 -2.232 was replaced to 165 -2.808 in same group
1 1.789 was replaced to 182 1.558 in same group
39 6.242 was replaced to 197 5.685 in same group
87 5.836 was replaced to 220 5.634 in same group
75 5.799 was replaced to 240 5.601 in same group
79 -1.358 was replaced to 245 -2.389 in same group
Bank No.: Energy
Bank 1: -17.014
Bank 2: -8.816
Bank 3: -11.490
Bank 4: 23.381
Bank 5: -18.273
Bank 6: -13.300
Bank 7: -17.246
Bank 8: -15.261
Bank 9: -6.440
Bank 10: -11.074
Bank 11: -10.754
Bank 12: -14.387
Bank 13: -19.119
Bank 14: -17.096
Bank 15: -6.416
Bank 16: 0.329
Bank 17: -12.729
Bank 18: -17.627
Bank 19: -17.639
Bank 20: -12.513
Bank 21: -15.704
Bank 22: 29.302
Bank 23: -3.206
Bank 24: -6.850
Bank 25: 2.476
Bank 26: -18.131
Bank 27: 1.413
Bank 28: -2.509
Bank 29: -16.706
Bank 30: 104.432
Bank 31: -13.290
Bank 32: -0.261
Bank 33: -17.803
Bank 34: 3.828
Bank 35: -18.140
Bank 36: -18.292
Bank 37: -6.621
Bank 38: -14.644
Bank 39: -8.934
Bank 40: -15.019
Bank 41: -15.273
Bank 42: -10.572
Bank 43: -7.233
Bank 44: -7.125
Bank 45: -15.396
Bank 46: -2.847
Bank 47: -12.974
Bank 48: -12.722
Bank 49: 2.398
Bank 50: -9.384
Bank 51: -7.725
Bank 52: -19.312
Bank 53: -8.530
Bank 54: -18.657
Bank 55: -10.692
Bank 56: -13.383
Bank 57: -18.551
Bank 58: -12.285
Bank 59: -10.040
Bank 60: -5.368
Bank 61: -16.728
Bank 62: -9.283
Bank 63: -15.991
Bank 64: -9.407
Bank 65: -20.055
Bank 66: -10.232
Bank 67: -12.252
Bank 68: -9.073
Bank 69: -12.832
Bank 70: -17.445
Bank 71: -10.848
Bank 72: -19.086
Bank 73: -18.355
Bank 74: -11.687
Bank 75: -13.235
Bank 76: -8.720
Bank 77: -16.693
Bank 78: -4.390
Bank 79: -10.924
Bank 80: -13.944
Bank 81: -16.000
Bank 82: -11.243
Bank 83: -6.668
Bank 84: -15.318
Bank 85: -5.422
Bank 86: -12.399
Bank 87: -14.302
Bank 88: -15.776
Bank 89: -17.945
Bank 90: -8.666
Bank 91: -19.070
Bank 92: -17.892
Bank 93: -13.682
Bank 94: -16.834
Bank 95: -18.945
Bank 96: -19.866
Bank 97: -18.377
Bank 98: -9.926
Bank 99: -17.427
Bank 100: -11.007
------------------------------------------------------------
CSA Run Number / Interation / SEED CYCLE = 1 / 1 / 84
D_ave / D_cut / D_min = 5.221 / 2.078 / 2.078
------------------------------------------------------------
Used time for CSA cycle 0D 1H 9M 6.9S
LCSA PLD: 1 1 20850 2.078 54 25 -18.657 2.476 5102044 45 100 4146.929 sec
------------------------------------------------------------
Select Seeds....
Unused seeds : 45 / 100
------------------------------------------------------------
Make new conformation
perturbation rate : 0.000
calc_ml_energy.mol2 is working!
Time for inference: 42.91 (s)
21 1.394 was replaced to 62 1.203 in same group
50 3.168 was replaced to 96 2.154 in same group
4 -0.758 was replaced to 142 -1.114 in same group
79 -2.389 was replaced to 200 -2.734 in same group
72 -1.785 was replaced to 211 -1.797 in same group
5 -2.808 was replaced to 224 -2.851 in same group
44 5.964 was replaced to 239 5.176 in same group
44 5.176 was replaced to 241 1.360 in same group
24 3.881 was replaced to 242 3.379 in same group
Bank No.: Energy
Bank 1: -17.014
Bank 2: -8.816
Bank 3: -11.490
Bank 4: -15.047
Bank 5: -18.309
Bank 6: -13.300
Bank 7: -17.246
Bank 8: -15.261
Bank 9: -6.440
Bank 10: -11.074
Bank 11: -10.754
Bank 12: -14.387
Bank 13: -19.119
Bank 14: -17.096
Bank 15: -6.416
Bank 16: 0.329
Bank 17: -12.729
Bank 18: -17.627
Bank 19: -17.639
Bank 20: -12.513
Bank 21: -13.922
Bank 22: 29.302
Bank 23: -3.206
Bank 24: -9.360
Bank 25: 2.476
Bank 26: -18.131
Bank 27: 1.413
Bank 28: -2.509
Bank 29: -16.706
Bank 30: 104.432
Bank 31: -13.290
Bank 32: -0.261
Bank 33: -17.803
Bank 34: 3.828
Bank 35: -18.140
Bank 36: -18.292
Bank 37: -6.621
Bank 38: -14.644
Bank 39: -8.934
Bank 40: -15.019
Bank 41: -15.273
Bank 42: -10.572
Bank 43: -7.233
Bank 44: -9.316
Bank 45: -15.396
Bank 46: -2.847
Bank 47: -12.974
Bank 48: -12.722
Bank 49: 2.398
Bank 50: -9.229
Bank 51: -7.725
Bank 52: -19.312
Bank 53: -8.530
Bank 54: -18.657
Bank 55: -10.692
Bank 56: -13.383
Bank 57: -18.551
Bank 58: -12.285
Bank 59: -10.040
Bank 60: -5.368
Bank 61: -16.728
Bank 62: -9.283
Bank 63: -15.991
Bank 64: -9.407
Bank 65: -20.055
Bank 66: -10.232
Bank 67: -12.252
Bank 68: -9.073
Bank 69: -12.832
Bank 70: -17.445
Bank 71: -10.848
Bank 72: -19.213
Bank 73: -18.355
Bank 74: -11.687
Bank 75: -13.235
Bank 76: -8.720
Bank 77: -16.693
Bank 78: -4.390
Bank 79: -12.521
Bank 80: -13.944
Bank 81: -16.000
Bank 82: -11.243
Bank 83: -6.668
Bank 84: -15.318
Bank 85: -5.422
Bank 86: -12.399
Bank 87: -14.302
Bank 88: -15.776
Bank 89: -17.945
Bank 90: -8.666
Bank 91: -19.070
Bank 92: -17.892
Bank 93: -13.682
Bank 94: -16.834
Bank 95: -18.945
Bank 96: -19.866
Bank 97: -18.377
Bank 98: -9.926
Bank 99: -17.427
Bank 100: -11.007
------------------------------------------------------------
CSA Run Number / Interation / SEED CYCLE = 1 / 1 / 85
D_ave / D_cut / D_min = 5.223 / 2.078 / 2.078
------------------------------------------------------------
Used time for CSA cycle 0D 1H 9M 59.5S
LCSA PLD: 1 1 21100 2.078 54 25 -18.657 2.476 5163244 27 100 4199.531 sec
------------------------------------------------------------
Select Seeds....
Unused seeds : 27 / 100
------------------------------------------------------------
Make new conformation
perturbation rate : 0.000
calc_ml_energy.mol2 is working!
Time for inference: 42.9 (s)
57 -1.451 was replaced to 62 -1.548 in same group
15 4.795 was replaced to 79 3.236 in same group
24 3.379 was replaced to 182 2.513 in same group
37 5.344 was replaced to 189 5.087 in same group
44 1.360 was replaced to 197 -0.620 in same group
92 -5.314 was replaced to 216 -5.820 in same group
25 6.470 was replaced to 233 1.542 in new group
Bank No.: Energy
Bank 1: -17.014
Bank 2: -8.816
Bank 3: -11.490
Bank 4: -15.047
Bank 5: -18.309
Bank 6: -13.300
Bank 7: -17.246
Bank 8: -15.261
Bank 9: -6.440
Bank 10: -11.074
Bank 11: -10.754
Bank 12: -14.387
Bank 13: -19.119
Bank 14: -17.096
Bank 15: -6.753
Bank 16: 0.329
Bank 17: -12.729
Bank 18: -17.627
Bank 19: -17.639
Bank 20: -12.513
Bank 21: -13.922
Bank 22: 29.302
Bank 23: -3.206
Bank 24: -4.217
Bank 25: 12.393
Bank 26: -18.131
Bank 27: 1.413
Bank 28: -2.509
Bank 29: -16.706
Bank 30: 104.432
Bank 31: -13.290
Bank 32: -0.261
Bank 33: -17.803
Bank 34: 3.828
Bank 35: -18.140
Bank 36: -18.292
Bank 37: -6.852
Bank 38: -14.644
Bank 39: -8.934
Bank 40: -15.019
Bank 41: -15.273
Bank 42: -10.572
Bank 43: -7.233
Bank 44: -11.056
Bank 45: -15.396
Bank 46: -2.847
Bank 47: -12.974
Bank 48: -12.722
Bank 49: 2.398
Bank 50: -9.229
Bank 51: -7.725
Bank 52: -19.312
Bank 53: -8.530
Bank 54: -18.657
Bank 55: -10.692
Bank 56: -13.383
Bank 57: -17.832
Bank 58: -12.285
Bank 59: -10.040
Bank 60: -5.368
Bank 61: -16.728
Bank 62: -9.283
Bank 63: -15.991
Bank 64: -9.407
Bank 65: -20.055
Bank 66: -10.232
Bank 67: -12.252
Bank 68: -9.073
Bank 69: -12.832
Bank 70: -17.445
Bank 71: -10.848
Bank 72: -19.213
Bank 73: -18.355
Bank 74: -11.687
Bank 75: -13.235
Bank 76: -8.720
Bank 77: -16.693
Bank 78: -4.390
Bank 79: -12.521
Bank 80: -13.944
Bank 81: -16.000
Bank 82: -11.243
Bank 83: -6.668
Bank 84: -15.318
Bank 85: -5.422
Bank 86: -12.399
Bank 87: -14.302
Bank 88: -15.776
Bank 89: -17.945
Bank 90: -8.666
Bank 91: -19.070
Bank 92: -18.009
Bank 93: -13.682
Bank 94: -16.834
Bank 95: -18.945
Bank 96: -19.866
Bank 97: -18.377
Bank 98: -9.926
Bank 99: -17.427
Bank 100: -11.007
------------------------------------------------------------
CSA Run Number / Interation / SEED CYCLE = 1 / 1 / 86
D_ave / D_cut / D_min = 5.196 / 2.078 / 2.078
------------------------------------------------------------
Used time for CSA cycle 0D 1H 10M 50.7S
LCSA PLD: 1 1 21350 2.078 92 30 -18.009 104.432 5222936 9 100 4250.724 sec
------------------------------------------------------------
Select Seeds....
Unused seeds : 9 / 100
------------------------------------------------------------
Make new conformation
perturbation rate : 0.000
calc_ml_energy.mol2 is working!
Time for inference: 44.51 (s)
30 6.461 was replaced to 75 6.226 in new group
96 -2.656 was replaced to 106 -3.325 in same group
15 3.236 was replaced to 177 3.010 in same group
3 6.391 was replaced to 183 6.256 in new group
57 -1.548 was replaced to 197 -1.610 in same group
4 -1.114 was replaced to 234 -1.250 in same group
Bank No.: Energy
Bank 1: -17.014
Bank 2: -8.816
Bank 3: -4.294
Bank 4: -14.654
Bank 5: -18.309
Bank 6: -13.300
Bank 7: -17.246
Bank 8: -15.261
Bank 9: -6.440
Bank 10: -11.074
Bank 11: -10.754
Bank 12: -14.387
Bank 13: -19.119
Bank 14: -17.096
Bank 15: -10.893
Bank 16: 0.329
Bank 17: -12.729
Bank 18: -17.627
Bank 19: -17.639
Bank 20: -12.513
Bank 21: -13.922
Bank 22: 29.302
Bank 23: -3.206
Bank 24: -4.217
Bank 25: 12.393
Bank 26: -18.131
Bank 27: 1.413
Bank 28: -2.509
Bank 29: -16.706
Bank 30: -7.625
Bank 31: -13.290
Bank 32: -0.261
Bank 33: -17.803
Bank 34: 3.828
Bank 35: -18.140
Bank 36: -18.292
Bank 37: -6.852
Bank 38: -14.644
Bank 39: -8.934
Bank 40: -15.019
Bank 41: -15.273
Bank 42: -10.572
Bank 43: -7.233
Bank 44: -11.056
Bank 45: -15.396
Bank 46: -2.847
Bank 47: -12.974
Bank 48: -12.722
Bank 49: 2.398
Bank 50: -9.229
Bank 51: -7.725
Bank 52: -19.312
Bank 53: -8.530
Bank 54: -18.657
Bank 55: -10.692
Bank 56: -13.383
Bank 57: -17.642
Bank 58: -12.285
Bank 59: -10.040
Bank 60: -5.368
Bank 61: -16.728
Bank 62: -9.283
Bank 63: -15.991
Bank 64: -9.407
Bank 65: -20.055
Bank 66: -10.232
Bank 67: -12.252
Bank 68: -9.073
Bank 69: -12.832
Bank 70: -17.445
Bank 71: -10.848
Bank 72: -19.213
Bank 73: -18.355
Bank 74: -11.687
Bank 75: -13.235
Bank 76: -8.720
Bank 77: -16.693
Bank 78: -4.390
Bank 79: -12.521
Bank 80: -13.944
Bank 81: -16.000
Bank 82: -11.243
Bank 83: -6.668
Bank 84: -15.318
Bank 85: -5.422
Bank 86: -12.399
Bank 87: -14.302
Bank 88: -15.776
Bank 89: -17.945
Bank 90: -8.666
Bank 91: -19.070
Bank 92: -18.009
Bank 93: -13.682
Bank 94: -16.834
Bank 95: -18.945
Bank 96: -18.798
Bank 97: -18.377
Bank 98: -9.926
Bank 99: -17.427
Bank 100: -11.007
------------------------------------------------------------
CSA Run Number / Interation / SEED CYCLE = 1 / 1 / 87
D_ave / D_cut / D_min = 5.184 / 2.078 / 2.078
------------------------------------------------------------
Used time for CSA cycle 0D 1H 11M 42.4S
LCSA PLD: 1 1 21600 2.078 92 3 -18.009 -4.294 5283323 6 100 4302.402 sec
------------------------------------------------------------
Select Seeds....
Unused seeds : 6 / 100
------------------------------------------------------------
Make new conformation
perturbation rate : 0.000
calc_ml_energy.mol2 is working!
Time for inference: 43.86 (s)
15 3.010 was replaced to 58 2.047 in same group
5 -2.851 was replaced to 122 -3.084 in same group
26 0.680 was replaced to 158 0.211 in same group
Bank No.: Energy
Bank 1: -17.014
Bank 2: -8.816
Bank 3: -4.294
Bank 4: -14.654
Bank 5: -18.232
Bank 6: -13.300
Bank 7: -17.246
Bank 8: -15.261
Bank 9: -6.440
Bank 10: -11.074
Bank 11: -10.754
Bank 12: -14.387
Bank 13: -19.119
Bank 14: -17.096
Bank 15: -9.588
Bank 16: 0.329
Bank 17: -12.729
Bank 18: -17.627
Bank 19: -17.639
Bank 20: -12.513
Bank 21: -13.922
Bank 22: 29.302
Bank 23: -3.206
Bank 24: -4.217
Bank 25: 12.393
Bank 26: -18.077
Bank 27: 1.413
Bank 28: -2.509
Bank 29: -16.706
Bank 30: -7.625
Bank 31: -13.290
Bank 32: -0.261
Bank 33: -17.803
Bank 34: 3.828
Bank 35: -18.140
Bank 36: -18.292
Bank 37: -6.852
Bank 38: -14.644
Bank 39: -8.934
Bank 40: -15.019
Bank 41: -15.273
Bank 42: -10.572
Bank 43: -7.233
Bank 44: -11.056
Bank 45: -15.396
Bank 46: -2.847
Bank 47: -12.974
Bank 48: -12.722
Bank 49: 2.398
Bank 50: -9.229
Bank 51: -7.725
Bank 52: -19.312
Bank 53: -8.530
Bank 54: -18.657
Bank 55: -10.692
Bank 56: -13.383
Bank 57: -17.642
Bank 58: -12.285
Bank 59: -10.040
Bank 60: -5.368
Bank 61: -16.728
Bank 62: -9.283
Bank 63: -15.991
Bank 64: -9.407
Bank 65: -20.055
Bank 66: -10.232
Bank 67: -12.252
Bank 68: -9.073
Bank 69: -12.832
Bank 70: -17.445
Bank 71: -10.848
Bank 72: -19.213
Bank 73: -18.355
Bank 74: -11.687
Bank 75: -13.235
Bank 76: -8.720
Bank 77: -16.693
Bank 78: -4.390
Bank 79: -12.521
Bank 80: -13.944
Bank 81: -16.000
Bank 82: -11.243
Bank 83: -6.668
Bank 84: -15.318
Bank 85: -5.422
Bank 86: -12.399
Bank 87: -14.302
Bank 88: -15.776
Bank 89: -17.945
Bank 90: -8.666
Bank 91: -19.070
Bank 92: -18.009
Bank 93: -13.682
Bank 94: -16.834
Bank 95: -18.945
Bank 96: -18.798
Bank 97: -18.377
Bank 98: -9.926
Bank 99: -17.427
Bank 100: -11.007
------------------------------------------------------------
CSA Run Number / Interation / SEED CYCLE = 1 / 1 / 88
D_ave / D_cut / D_min = 5.178 / 2.078 / 2.078
------------------------------------------------------------
Used time for CSA cycle 0D 1H 12M 33.5S
LCSA PLD: 1 1 21850 2.078 92 3 -18.009 -4.294 5341852 3 100 4353.469 sec
------------------------------------------------------------
Select Seeds....
Unused seeds : 3 / 100
------------------------------------------------------------
Make new conformation
perturbation rate : 0.000
calc_ml_energy.mol2 is working!
Time for inference: 43.1 (s)
41 3.756 was replaced to 18 3.184 in same group
15 2.047 was replaced to 54 1.794 in same group
83 5.822 was replaced to 128 5.246 in same group
21 1.203 was replaced to 131 0.963 in same group
15 1.794 was replaced to 180 1.730 in same group
15 1.730 was replaced to 181 1.406 in same group
Bank No.: Energy
Bank 1: -17.014
Bank 2: -8.816
Bank 3: -4.294
Bank 4: -14.654
Bank 5: -18.232
Bank 6: -13.300
Bank 7: -17.246
Bank 8: -15.261
Bank 9: -6.440
Bank 10: -11.074
Bank 11: -10.754
Bank 12: -14.387
Bank 13: -19.119
Bank 14: -17.096
Bank 15: -5.022
Bank 16: 0.329
Bank 17: -12.729
Bank 18: -17.627
Bank 19: -17.639
Bank 20: -12.513
Bank 21: -16.924
Bank 22: 29.302
Bank 23: -3.206
Bank 24: -4.217
Bank 25: 12.393
Bank 26: -18.077
Bank 27: 1.413
Bank 28: -2.509
Bank 29: -16.706
Bank 30: -7.625
Bank 31: -13.290
Bank 32: -0.261
Bank 33: -17.803
Bank 34: 3.828
Bank 35: -18.140
Bank 36: -18.292
Bank 37: -6.852
Bank 38: -14.644
Bank 39: -8.934
Bank 40: -15.019
Bank 41: -9.727
Bank 42: -10.572
Bank 43: -7.233
Bank 44: -11.056
Bank 45: -15.396
Bank 46: -2.847
Bank 47: -12.974
Bank 48: -12.722
Bank 49: 2.398
Bank 50: -9.229
Bank 51: -7.725
Bank 52: -19.312
Bank 53: -8.530
Bank 54: -18.657
Bank 55: -10.692
Bank 56: -13.383
Bank 57: -17.642
Bank 58: -12.285
Bank 59: -10.040
Bank 60: -5.368
Bank 61: -16.728
Bank 62: -9.283
Bank 63: -15.991
Bank 64: -9.407
Bank 65: -20.055
Bank 66: -10.232
Bank 67: -12.252
Bank 68: -9.073
Bank 69: -12.832
Bank 70: -17.445
Bank 71: -10.848
Bank 72: -19.213
Bank 73: -18.355
Bank 74: -11.687
Bank 75: -13.235
Bank 76: -8.720
Bank 77: -16.693
Bank 78: -4.390
Bank 79: -12.521
Bank 80: -13.944
Bank 81: -16.000
Bank 82: -11.243
Bank 83: -6.698
Bank 84: -15.318
Bank 85: -5.422
Bank 86: -12.399
Bank 87: -14.302
Bank 88: -15.776
Bank 89: -17.945
Bank 90: -8.666
Bank 91: -19.070
Bank 92: -18.009
Bank 93: -13.682
Bank 94: -16.834
Bank 95: -18.945
Bank 96: -18.798
Bank 97: -18.377
Bank 98: -9.926
Bank 99: -17.427
Bank 100: -11.007
------------------------------------------------------------
CSA Run Number / Interation / SEED CYCLE = 1 / 1 / 89
D_ave / D_cut / D_min = 5.172 / 2.078 / 2.078
------------------------------------------------------------
Used time for CSA cycle 0D 1H 13M 25.0S
LCSA PLD: 1 1 22100 2.078 92 3 -18.009 -4.294 5402476 4 100 4404.995 sec
------------------------------------------------------------
Select Seeds....
Unused seeds : 4 / 100
------------------------------------------------------------
Make new conformation
perturbation rate : 0.000
calc_ml_energy.mol2 is working!
Time for inference: 43.41 (s)
6 4.561 was replaced to 108 4.160 in same group
83 5.246 was replaced to 132 4.407 in same group
15 1.406 was replaced to 176 0.820 in same group
36 -4.004 was replaced to 194 -4.404 in same group
Bank No.: Energy
Bank 1: -17.014
Bank 2: -8.816
Bank 3: -4.294
Bank 4: -14.654
Bank 5: -18.232
Bank 6: -11.644
Bank 7: -17.246
Bank 8: -15.261
Bank 9: -6.440
Bank 10: -11.074
Bank 11: -10.754
Bank 12: -14.387
Bank 13: -19.119
Bank 14: -17.096
Bank 15: -14.556
Bank 16: 0.329
Bank 17: -12.729
Bank 18: -17.627
Bank 19: -17.639
Bank 20: -12.513
Bank 21: -16.924
Bank 22: 29.302
Bank 23: -3.206
Bank 24: -4.217
Bank 25: 12.393
Bank 26: -18.077
Bank 27: 1.413
Bank 28: -2.509
Bank 29: -16.706
Bank 30: -7.625
Bank 31: -13.290
Bank 32: -0.261
Bank 33: -17.803
Bank 34: 3.828
Bank 35: -18.140
Bank 36: -17.172
Bank 37: -6.852
Bank 38: -14.644
Bank 39: -8.934
Bank 40: -15.019
Bank 41: -9.727
Bank 42: -10.572
Bank 43: -7.233
Bank 44: -11.056
Bank 45: -15.396
Bank 46: -2.847
Bank 47: -12.974
Bank 48: -12.722
Bank 49: 2.398
Bank 50: -9.229
Bank 51: -7.725
Bank 52: -19.312
Bank 53: -8.530
Bank 54: -18.657
Bank 55: -10.692
Bank 56: -13.383
Bank 57: -17.642
Bank 58: -12.285
Bank 59: -10.040
Bank 60: -5.368
Bank 61: -16.728
Bank 62: -9.283
Bank 63: -15.991
Bank 64: -9.407
Bank 65: -20.055
Bank 66: -10.232
Bank 67: -12.252
Bank 68: -9.073
Bank 69: -12.832
Bank 70: -17.445
Bank 71: -10.848
Bank 72: -19.213
Bank 73: -18.355
Bank 74: -11.687
Bank 75: -13.235
Bank 76: -8.720
Bank 77: -16.693
Bank 78: -4.390
Bank 79: -12.521
Bank 80: -13.944
Bank 81: -16.000
Bank 82: -11.243
Bank 83: -9.197
Bank 84: -15.318
Bank 85: -5.422
Bank 86: -12.399
Bank 87: -14.302
Bank 88: -15.776
Bank 89: -17.945
Bank 90: -8.666
Bank 91: -19.070
Bank 92: -18.009
Bank 93: -13.682
Bank 94: -16.834
Bank 95: -18.945
Bank 96: -18.798
Bank 97: -18.377
Bank 98: -9.926
Bank 99: -17.427
Bank 100: -11.007
------------------------------------------------------------
CSA Run Number / Interation / SEED CYCLE = 1 / 1 / 90
D_ave / D_cut / D_min = 5.169 / 2.078 / 2.078
------------------------------------------------------------
Used time for CSA cycle 0D 1H 14M 18.7S
LCSA PLD: 1 1 22350 2.078 92 3 -18.009 -4.294 5469705 4 100 4458.714 sec
------------------------------------------------------------
Select Seeds....
Unused seeds : 4 / 100
------------------------------------------------------------
Make new conformation
perturbation rate : 0.000
calc_ml_energy.mol2 is working!
Time for inference: 44.54 (s)
15 0.820 was replaced to 128 0.561 in same group
15 0.561 was replaced to 129 0.453 in same group
15 0.453 was replaced to 179 -0.453 in same group
83 4.407 was replaced to 185 4.043 in same group
83 4.043 was replaced to 186 2.787 in same group
4 -1.250 was replaced to 213 -1.377 in same group
99 0.718 was replaced to 218 0.333 in same group
18 1.190 was replaced to 233 1.122 in same group
18 1.122 was replaced to 234 0.446 in same group
Bank No.: Energy
Bank 1: -17.014
Bank 2: -8.816
Bank 3: -4.294
Bank 4: -14.846
Bank 5: -18.232
Bank 6: -11.644
Bank 7: -17.246
Bank 8: -15.261
Bank 9: -6.440
Bank 10: -11.074
Bank 11: -10.754
Bank 12: -14.387
Bank 13: -19.119
Bank 14: -17.096
Bank 15: -13.985
Bank 16: 0.329
Bank 17: -12.729
Bank 18: -18.696
Bank 19: -17.639
Bank 20: -12.513
Bank 21: -16.924
Bank 22: 29.302
Bank 23: -3.206
Bank 24: -4.217
Bank 25: 12.393
Bank 26: -18.077
Bank 27: 1.413
Bank 28: -2.509
Bank 29: -16.706
Bank 30: -7.625
Bank 31: -13.290
Bank 32: -0.261
Bank 33: -17.803
Bank 34: 3.828
Bank 35: -18.140
Bank 36: -17.172
Bank 37: -6.852
Bank 38: -14.644
Bank 39: -8.934
Bank 40: -15.019
Bank 41: -9.727
Bank 42: -10.572
Bank 43: -7.233
Bank 44: -11.056
Bank 45: -15.396
Bank 46: -2.847
Bank 47: -12.974
Bank 48: -12.722
Bank 49: 2.398
Bank 50: -9.229
Bank 51: -7.725
Bank 52: -19.312
Bank 53: -8.530
Bank 54: -18.657
Bank 55: -10.692
Bank 56: -13.383
Bank 57: -17.642
Bank 58: -12.285
Bank 59: -10.040
Bank 60: -5.368
Bank 61: -16.728
Bank 62: -9.283
Bank 63: -15.991
Bank 64: -9.407
Bank 65: -20.055
Bank 66: -10.232
Bank 67: -12.252
Bank 68: -9.073
Bank 69: -12.832
Bank 70: -17.445
Bank 71: -10.848
Bank 72: -19.213
Bank 73: -18.355
Bank 74: -11.687
Bank 75: -13.235
Bank 76: -8.720
Bank 77: -16.693
Bank 78: -4.390
Bank 79: -12.521
Bank 80: -13.944
Bank 81: -16.000
Bank 82: -11.243
Bank 83: -8.668
Bank 84: -15.318
Bank 85: -5.422
Bank 86: -12.399
Bank 87: -14.302
Bank 88: -15.776
Bank 89: -17.945
Bank 90: -8.666
Bank 91: -19.070
Bank 92: -18.009
Bank 93: -13.682
Bank 94: -16.834
Bank 95: -18.945
Bank 96: -18.798
Bank 97: -18.377
Bank 98: -9.926
Bank 99: -17.979
Bank 100: -11.007
------------------------------------------------------------
CSA Run Number / Interation / SEED CYCLE = 1 / 1 / 91
D_ave / D_cut / D_min = 5.176 / 2.078 / 2.078
------------------------------------------------------------
Used time for CSA cycle 0D 1H 15M 11.9S
LCSA PLD: 1 1 22600 2.078 92 3 -18.009 -4.294 5534604 5 100 4511.896 sec
------------------------------------------------------------
Select Seeds....
Unused seeds : 5 / 100
------------------------------------------------------------
Make new conformation
perturbation rate : 0.000
calc_ml_energy.mol2 is working!
Time for inference: 43.7 (s)
23 4.266 was replaced to 56 2.847 in same group
2 3.457 was replaced to 82 2.517 in same group
4 -1.377 was replaced to 126 -1.595 in same group
12 3.240 was replaced to 152 3.109 in same group
15 -0.453 was replaced to 181 -1.211 in same group
34 3.893 was replaced to 209 3.731 in same group
47 3.563 was replaced to 236 3.558 in same group
52 -2.183 was replaced to 249 -3.174 in same group
Bank No.: Energy
Bank 1: -17.014
Bank 2: 2.579
Bank 3: -4.294
Bank 4: -13.591
Bank 5: -18.232
Bank 6: -11.644
Bank 7: -17.246
Bank 8: -15.261
Bank 9: -6.440
Bank 10: -11.074
Bank 11: -10.754
Bank 12: -11.180
Bank 13: -19.119
Bank 14: -17.096
Bank 15: -13.445
Bank 16: 0.329
Bank 17: -12.729
Bank 18: -18.696
Bank 19: -17.639
Bank 20: -12.513
Bank 21: -16.924
Bank 22: 29.302
Bank 23: -13.785
Bank 24: -4.217
Bank 25: 12.393
Bank 26: -18.077
Bank 27: 1.413
Bank 28: -2.509
Bank 29: -16.706
Bank 30: -7.625
Bank 31: -13.290
Bank 32: -0.261
Bank 33: -17.803
Bank 34: -19.197
Bank 35: -18.140
Bank 36: -17.172
Bank 37: -6.852
Bank 38: -14.644
Bank 39: -8.934
Bank 40: -15.019
Bank 41: -9.727
Bank 42: -10.572
Bank 43: -7.233
Bank 44: -11.056
Bank 45: -15.396
Bank 46: -2.847
Bank 47: -12.909
Bank 48: -12.722
Bank 49: 2.398
Bank 50: -9.229
Bank 51: -7.725
Bank 52: -20.989
Bank 53: -8.530
Bank 54: -18.657
Bank 55: -10.692
Bank 56: -13.383
Bank 57: -17.642
Bank 58: -12.285
Bank 59: -10.040
Bank 60: -5.368
Bank 61: -16.728
Bank 62: -9.283
Bank 63: -15.991
Bank 64: -9.407
Bank 65: -20.055
Bank 66: -10.232
Bank 67: -12.252
Bank 68: -9.073
Bank 69: -12.832
Bank 70: -17.445
Bank 71: -10.848
Bank 72: -19.213
Bank 73: -18.355
Bank 74: -11.687
Bank 75: -13.235
Bank 76: -8.720
Bank 77: -16.693
Bank 78: -4.390
Bank 79: -12.521
Bank 80: -13.944
Bank 81: -16.000
Bank 82: -11.243
Bank 83: -8.668
Bank 84: -15.318
Bank 85: -5.422
Bank 86: -12.399
Bank 87: -14.302
Bank 88: -15.776
Bank 89: -17.945
Bank 90: -8.666
Bank 91: -19.070
Bank 92: -18.009
Bank 93: -13.682
Bank 94: -16.834
Bank 95: -18.945
Bank 96: -18.798
Bank 97: -18.377
Bank 98: -9.926
Bank 99: -17.979
Bank 100: -11.007
------------------------------------------------------------
CSA Run Number / Interation / SEED CYCLE = 1 / 1 / 92
D_ave / D_cut / D_min = 5.163 / 2.078 / 2.078
------------------------------------------------------------
Used time for CSA cycle 0D 1H 16M 3.2S
LCSA PLD: 1 1 22850 2.078 92 3 -18.009 -4.294 5597596 8 100 4563.150 sec
------------------------------------------------------------
Select Seeds....
Unused seeds : 8 / 100
------------------------------------------------------------
Make new conformation
perturbation rate : 0.000
calc_ml_energy.mol2 is working!
Time for inference: 42.4 (s)
35 -0.014 was replaced to 90 -0.294 in same group
47 3.558 was replaced to 138 3.422 in same group
99 0.333 was replaced to 152 0.044 in same group
38 5.565 was replaced to 157 5.516 in same group
83 2.787 was replaced to 185 0.944 in same group
70 0.032 was replaced to 218 -0.368 in same group
7 -1.271 was replaced to 236 -1.487 in same group
88 -3.344 was replaced to 243 -3.745 in same group
Bank No.: Energy
Bank 1: -17.014
Bank 2: 2.579
Bank 3: -4.294
Bank 4: -13.591
Bank 5: -18.232
Bank 6: -11.644
Bank 7: -15.626
Bank 8: -15.261
Bank 9: -6.440
Bank 10: -11.074
Bank 11: -10.754
Bank 12: -11.180
Bank 13: -19.119
Bank 14: -17.096
Bank 15: -13.445
Bank 16: 0.329
Bank 17: -12.729
Bank 18: -18.696
Bank 19: -17.639
Bank 20: -12.513
Bank 21: -16.924
Bank 22: 29.302
Bank 23: -13.785
Bank 24: -4.217
Bank 25: 12.393
Bank 26: -18.077
Bank 27: 1.413
Bank 28: -2.509
Bank 29: -16.706
Bank 30: -7.625
Bank 31: -13.290
Bank 32: -0.261
Bank 33: -17.803
Bank 34: -19.197
Bank 35: -17.837
Bank 36: -17.172
Bank 37: -6.852
Bank 38: -16.881
Bank 39: -8.934
Bank 40: -15.019
Bank 41: -9.727
Bank 42: -10.572
Bank 43: -7.233
Bank 44: -11.056
Bank 45: -15.396
Bank 46: -2.847
Bank 47: -11.853
Bank 48: -12.722
Bank 49: 2.398
Bank 50: -9.229
Bank 51: -7.725
Bank 52: -20.989
Bank 53: -8.530
Bank 54: -18.657
Bank 55: -10.692
Bank 56: -13.383
Bank 57: -17.642
Bank 58: -12.285
Bank 59: -10.040
Bank 60: -5.368
Bank 61: -16.728
Bank 62: -9.283
Bank 63: -15.991
Bank 64: -9.407
Bank 65: -20.055
Bank 66: -10.232
Bank 67: -12.252
Bank 68: -9.073
Bank 69: -12.832
Bank 70: -16.065
Bank 71: -10.848
Bank 72: -19.213
Bank 73: -18.355
Bank 74: -11.687
Bank 75: -13.235
Bank 76: -8.720
Bank 77: -16.693
Bank 78: -4.390
Bank 79: -12.521
Bank 80: -13.944
Bank 81: -16.000
Bank 82: -11.243
Bank 83: -11.019
Bank 84: -15.318
Bank 85: -5.422
Bank 86: -12.399
Bank 87: -14.302
Bank 88: -7.290
Bank 89: -17.945
Bank 90: -8.666
Bank 91: -19.070
Bank 92: -18.009
Bank 93: -13.682
Bank 94: -16.834
Bank 95: -18.945
Bank 96: -18.798
Bank 97: -18.377
Bank 98: -9.926
Bank 99: -15.804
Bank 100: -11.007
------------------------------------------------------------
CSA Run Number / Interation / SEED CYCLE = 1 / 1 / 93
D_ave / D_cut / D_min = 5.155 / 2.078 / 2.078
------------------------------------------------------------
Used time for CSA cycle 0D 1H 16M 53.8S
LCSA PLD: 1 1 23100 2.078 92 3 -18.009 -4.294 5664463 8 100 4613.770 sec
------------------------------------------------------------
Select Seeds....
Unused seeds : 8 / 100
------------------------------------------------------------
Make new conformation
perturbation rate : 0.000
calc_ml_energy.mol2 is working!
Time for inference: 41.26 (s)
35 -0.294 was replaced to 4 -0.633 in same group
10 2.234 was replaced to 107 1.497 in same group
83 0.944 was replaced to 136 -0.103 in same group
35 -0.633 was replaced to 181 -1.138 in same group
70 -0.368 was replaced to 190 -0.534 in same group
87 5.634 was replaced to 221 5.430 in same group
50 2.154 was replaced to 230 1.614 in same group
65 -5.023 was replaced to 240 -5.344 in same group
Bank No.: Energy
Bank 1: -17.014
Bank 2: 2.579
Bank 3: -4.294
Bank 4: -13.591
Bank 5: -18.232
Bank 6: -11.644
Bank 7: -15.626
Bank 8: -15.261
Bank 9: -6.440
Bank 10: -9.210
Bank 11: -10.754
Bank 12: -11.180
Bank 13: -19.119
Bank 14: -17.096
Bank 15: -13.445
Bank 16: 0.329
Bank 17: -12.729
Bank 18: -18.696
Bank 19: -17.639
Bank 20: -12.513
Bank 21: -16.924
Bank 22: 29.302
Bank 23: -13.785
Bank 24: -4.217
Bank 25: 12.393
Bank 26: -18.077
Bank 27: 1.413
Bank 28: -2.509
Bank 29: -16.706
Bank 30: -7.625
Bank 31: -13.290
Bank 32: -0.261
Bank 33: -17.803
Bank 34: -19.197
Bank 35: -18.767
Bank 36: -17.172
Bank 37: -6.852
Bank 38: -16.881
Bank 39: -8.934
Bank 40: -15.019
Bank 41: -9.727
Bank 42: -10.572
Bank 43: -7.233
Bank 44: -11.056
Bank 45: -15.396
Bank 46: -2.847
Bank 47: -11.853
Bank 48: -12.722
Bank 49: 2.398
Bank 50: -8.327
Bank 51: -7.725
Bank 52: -20.989
Bank 53: -8.530
Bank 54: -18.657
Bank 55: -10.692
Bank 56: -13.383
Bank 57: -17.642
Bank 58: -12.285
Bank 59: -10.040
Bank 60: -5.368
Bank 61: -16.728
Bank 62: -9.283
Bank 63: -15.991
Bank 64: -9.407
Bank 65: -19.106
Bank 66: -10.232
Bank 67: -12.252
Bank 68: -9.073
Bank 69: -12.832
Bank 70: -16.931
Bank 71: -10.848
Bank 72: -19.213
Bank 73: -18.355
Bank 74: -11.687
Bank 75: -13.235
Bank 76: -8.720
Bank 77: -16.693
Bank 78: -4.390
Bank 79: -12.521
Bank 80: -13.944
Bank 81: -16.000
Bank 82: -11.243
Bank 83: -12.325
Bank 84: -15.318
Bank 85: -5.422
Bank 86: -12.399
Bank 87: -14.867
Bank 88: -7.290
Bank 89: -17.945
Bank 90: -8.666
Bank 91: -19.070
Bank 92: -18.009
Bank 93: -13.682
Bank 94: -16.834
Bank 95: -18.945
Bank 96: -18.798
Bank 97: -18.377
Bank 98: -9.926
Bank 99: -15.804
Bank 100: -11.007
------------------------------------------------------------
CSA Run Number / Interation / SEED CYCLE = 1 / 1 / 94
D_ave / D_cut / D_min = 5.154 / 2.078 / 2.078
------------------------------------------------------------
Used time for CSA cycle 0D 1H 17M 44.1S
LCSA PLD: 1 1 23350 2.078 92 3 -18.009 -4.294 5728191 7 100 4664.068 sec
------------------------------------------------------------
Select Seeds....
Unused seeds : 7 / 100
------------------------------------------------------------
Make new conformation
perturbation rate : 0.000
calc_ml_energy.mol2 is working!
Time for inference: 40.13 (s)
23 2.847 was replaced to 107 2.464 in same group
34 3.731 was replaced to 158 3.403 in same group
Bank No.: Energy
Bank 1: -17.014
Bank 2: 2.579
Bank 3: -4.294
Bank 4: -13.591
Bank 5: -18.232
Bank 6: -11.644
Bank 7: -15.626
Bank 8: -15.261
Bank 9: -6.440
Bank 10: -9.210
Bank 11: -10.754
Bank 12: -11.180
Bank 13: -19.119
Bank 14: -17.096
Bank 15: -13.445
Bank 16: 0.329
Bank 17: -12.729
Bank 18: -18.696
Bank 19: -17.639
Bank 20: -12.513
Bank 21: -16.924
Bank 22: 29.302
Bank 23: -12.384
Bank 24: -4.217
Bank 25: 12.393
Bank 26: -18.077
Bank 27: 1.413
Bank 28: -2.509
Bank 29: -16.706
Bank 30: -7.625
Bank 31: -13.290
Bank 32: -0.261
Bank 33: -17.803
Bank 34: -19.439
Bank 35: -18.767
Bank 36: -17.172
Bank 37: -6.852
Bank 38: -16.881
Bank 39: -8.934
Bank 40: -15.019
Bank 41: -9.727
Bank 42: -10.572
Bank 43: -7.233
Bank 44: -11.056
Bank 45: -15.396
Bank 46: -2.847
Bank 47: -11.853
Bank 48: -12.722
Bank 49: 2.398
Bank 50: -8.327
Bank 51: -7.725
Bank 52: -20.989
Bank 53: -8.530
Bank 54: -18.657
Bank 55: -10.692
Bank 56: -13.383
Bank 57: -17.642
Bank 58: -12.285
Bank 59: -10.040
Bank 60: -5.368
Bank 61: -16.728
Bank 62: -9.283
Bank 63: -15.991
Bank 64: -9.407
Bank 65: -19.106
Bank 66: -10.232
Bank 67: -12.252
Bank 68: -9.073
Bank 69: -12.832
Bank 70: -16.931
Bank 71: -10.848
Bank 72: -19.213
Bank 73: -18.355
Bank 74: -11.687
Bank 75: -13.235
Bank 76: -8.720
Bank 77: -16.693
Bank 78: -4.390
Bank 79: -12.521
Bank 80: -13.944
Bank 81: -16.000
Bank 82: -11.243
Bank 83: -12.325
Bank 84: -15.318
Bank 85: -5.422
Bank 86: -12.399
Bank 87: -14.867
Bank 88: -7.290
Bank 89: -17.945
Bank 90: -8.666
Bank 91: -19.070
Bank 92: -18.009
Bank 93: -13.682
Bank 94: -16.834
Bank 95: -18.945
Bank 96: -18.798
Bank 97: -18.377
Bank 98: -9.926
Bank 99: -15.804
Bank 100: -11.007
calc_ml_energy.mol2 is working!
Time for inference: 24.49 (s)
------------------------------------------------------------
Total User Time = 4798.725 sec
0 dy 1 hr 19 min 58.725 sec
------------------------------------------------------------
Parameter files exist
Error: forrtl: warning (768): Internal file write-to-self; undefined results
forrtl: warning (768): Internal file write-to-self; undefined results
forrtl: warning (768): Internal file write-to-self; undefined results
forrtl: warning (768): Internal file write-to-self; undefined results
forrtl: warning (768): Internal file write-to-self; undefined results
forrtl: warning (768): Internal file write-to-self; undefined results
forrtl: warning (768): Internal file write-to-self; undefined results
forrtl: warning (768): Internal file write-to-self; undefined results
forrtl: warning (768): Internal file write-to-self; undefined results
forrtl: warning (768): Internal file write-to-self; undefined results
GDDL outputs the top 50 protein-ligand docking poses. By selecting a ranking, you can visualize the corresponding docking pose. The output from GDDL shows the protein and ligand in rainbow and gray, respectively, while the crystal structure’s protein and ligand are displayed in pink and yellow. Depending on whether the show_crystal_structure option is checked, the crystal structure may or may not be displayed.
[ ]:
#@title Visualizing Protein-ligand Docking Output Files with the Crystal Structure
rank = 15 #@param {type:"integer"}
show_crystal_structure = True #@param {type:"boolean"}
wrt = True
write = []
if whether_covalent_docking_or_not == 'non-covalent':
fname = 'GalaxyDock_fb.mol2'
else:
fname = 'reranked_fb.mol2'
num = 0
with open('GalaxyDock_fb.E.info') as fp:
for line in fp:
num += 1
if num > 3 and num == rank+3:
score = line.split()[1].strip('\n')
break
num = 0
with open(fname) as fp:
for line in fp:
if line == '@<TRIPOS>MOLECULE\n':
num += 1
if num > rank:
break
if num == rank:
write.append(line)
fout = open('final_ligand.mol2', 'wt')
fout.writelines(write)
fout.close()
view = py3Dmol.view(width=800, height=600)
mol = next(pybel.readfile("mol2", "final_ligand.mol2"))
pdb_data = mol.write("pdb")
view.addModel(pdb_data, "pdb")
view.setStyle({'model': 0}, {"stick":{}})
view.addModel(open(f'/content/iitp_demonstrate/{target}_pro.pdb').read(), "pdb")
view.setBackgroundColor("white")
view.addSurface(py3Dmol.VDW, {"opacity":0.7, 'color':'white'}, {'chain':'A'})
view.setStyle({'model': 1}, {'cartoon': {'color': 'spectrum'}})
if show_crystal_structure:
view.addModel(open(f'/content/iitp_demonstrate/{target}_lig.pdb').read(), "pdb")
view.setStyle({'model': 2}, {'stick': {'color': 'yellow'}})
view.addaddLabel(f"GDDL Score: {score}", {'position': (50, 50, 50), 'color': 'red', 'fontSize': 20, 'backgroundColor': 'white'})
view.zoomTo()
view.show()
print(f'GDDL Score: {score}')
3Dmol.js failed to load for some reason. Please check your browser console for error messages.
GDDL Score: 21.537
The following graph shows the GDDL energy score according to rank.
[ ]:
#@title Plotting docking scores
from pathlib import Path
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
# Load energy data
output_energy_file = Path('GalaxyDock_fb.E.info')
energy_array = np.array([float(line.split()[1]) for line in output_energy_file.read_text().splitlines() if not line.startswith('!') and not line.startswith('Bank')], dtype=np.float64)
rank_array = np.array(range(1, 51))
# Set font size multiplier for better readability
font_size = 2
# Seaborn style and theme setup
sns.set_style("whitegrid") # Clean background with grid
sns.set_theme(rc={'figure.figsize':(8, 6)}) # Adjust figure size for better aspect ratio
sns.set_theme(font_scale=1.5) # Moderate scaling for better font size balance
# Create rank range for the x-axis
rmsd_range = list(range(1, 52))
# Create the plot
fig, ax = plt.subplots() # Create figure and axis
# Scatter plot with adjusted point size and color
ax = sns.scatterplot(x=rank_array, y=energy_array, s=100, color='royalblue', edgecolor='black', alpha=0.7)
# Customize axis ticks
plt.xticks(fontsize=font_size*3, fontweight='bold') # Increase x-tick font size
plt.yticks(fontsize=font_size*3, fontweight='bold') # Increase y-tick font size
# Set axis labels with enhanced font sizes and label padding
plt.xlabel('Rank', fontsize=font_size*6.0, labelpad=font_size*2, fontweight='bold')
plt.ylabel('Score', fontsize=font_size*6.0, labelpad=font_size*2, fontweight='bold')
# Add a vertical line for visual separation (at rank 2)
ax.axvline(2.0, linestyle='--', color='black', linewidth=1.5)
# Customize axis spines and gridlines
ax.spines['bottom'].set_color('black')
ax.spines['top'].set_color('black')
ax.spines['right'].set_color('black')
ax.spines['left'].set_color('black')
# Customize x-ticks to match the rank range
ax.set_xticks(np.array(rmsd_range))
# Set plot title with a larger font size
ax.set_title("Rank vs Score", fontsize=font_size*7, fontweight='bold')
# Adjust layout to prevent clipping and enhance spacing
plt.tight_layout()
# Show the plot
plt.show()
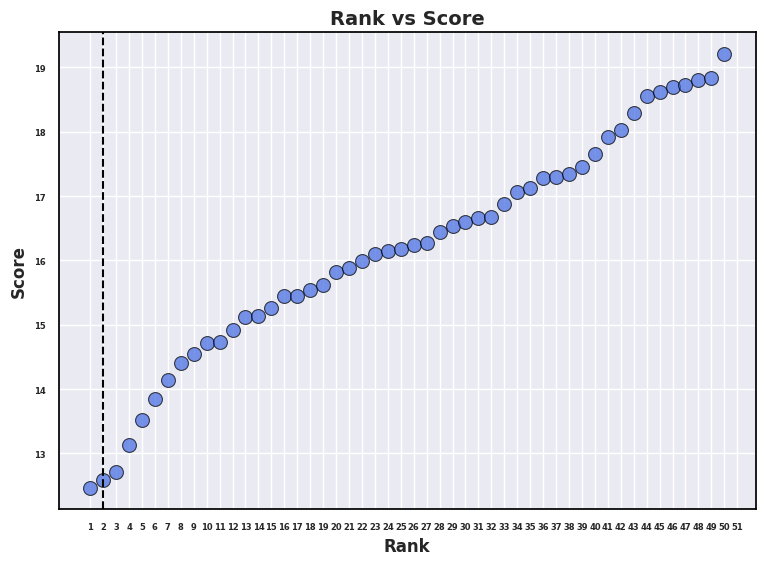
Calculate the RMSD between the crystal structure and the GDDL docking pose selected from the ranking above.
[ ]:
#@title Calculating RMSD of predicted structures
num = 0
with open(f'/content/iitp_demonstrate/{target}/rmsd.info') as fp:
for line in fp:
num += 1
if num == rank:
rmsd = line.split()[3].strip('\n')
print(f'RMSD between the crystal structure and the GDDL docking pose selected from the ranking above: {rmsd} Å')
---------------------------------------------------------------------------
FileNotFoundError Traceback (most recent call last)
<ipython-input-30-aaac157233ac> in <cell line: 4>()
2
3 num = 0
----> 4 with open(f'/content/iitp_demonstrate/{target}/rmsd.info') as fp:
5 for line in fp:
6 num += 1
FileNotFoundError: [Errno 2] No such file or directory: '/content/iitp_demonstrate/2qeh/rmsd.info'
GDDL does not output just a single structure; instead, it provides 50 different structures ranked by increasing energy. This can also be helpful in studying the multi-state of the target ligand. By examining the relationship between the actual GDDL energy and the RMSD of each docking pose, we can observe a significant correlation. In other words, the GDDL energy is indeed useful in assessing the stability of the structures.
[ ]:
#@title Plotting the relationship between RMSD and docking energy
import pandas as pd
import matplotlib.pyplot as plt
import numpy as np
import seaborn as sns
from scipy.stats import linregress, kendalltau, spearmanr
from sklearn.metrics import mean_absolute_error, mean_squared_error
from pathlib import Path
# Function to add trendline and display regression metrics
def add_trendline_and_metrics(ax, x, y, title):
slope, intercept, r_value, p_value, std_err = linregress(x, y)
trendline = np.array(x) * slope + intercept
ax.plot(x, trendline, color="g", linestyle="--", label=f"y = {slope:.2f}x + {intercept:.2f}")
# Calculating Kendall Tau and Spearman correlation
tau, _ = kendalltau(x, y)
spearman_corr, _ = spearmanr(x, y)
# Calculating MAE and RMSE
mae = mean_absolute_error(y, np.array(x) * slope + intercept)
rmse = np.sqrt(mean_squared_error(y, np.array(x) * slope + intercept))
# Display metrics on the plot with adjusted font size
ax.text(
0.05, 0.85,
f"$R^2$ = {r_value**2:.2f}\n"
f"p = {p_value:.2e}\n"
f"Kendall Tau = {tau:.2f}\n"
f"Spearman = {spearman_corr:.2f}\n"
f"MAE = {mae:.2f}\n"
f"RMSE = {rmse:.2f}",
transform=ax.transAxes,
fontsize=12, # Adjusted font size for metrics
verticalalignment="top",
)
ax.legend(loc="upper left", fontsize=12) # Adjusted font size for legend
# Load RMSD and energy data
rmsd_results = energy_array = np.array([float(line.split()[3].strip('\n')) for line in Path('rmsd.info').read_text().splitlines() if not line.startswith('!') and not line.startswith('Bank')], dtype=np.float64)
energy_array = np.array([float(line.split()[1]) for line in Path('GalaxyDock_fb.E.info').read_text().splitlines() if not line.startswith('!') and not line.startswith('Bank')], dtype=np.float64)
# Create a scatter plot using Seaborn
sns.set(style="whitegrid") # Set Seaborn style
fig, ax = plt.subplots(figsize=(8, 6)) # Create figure and axis
sns.scatterplot(x=rmsd_results, y=energy_array, ax=ax, s=50, color='b', alpha=0.5, edgecolors='k')
# Add trendline and metrics
add_trendline_and_metrics(ax, rmsd_results, energy_array, "RMSD vs GDDL Score")
# Set titles and labels with adjusted font sizes
ax.set_title("RMSD vs GDDL Score", fontsize=16)
ax.set_xlabel("RMSD", fontsize=14)
ax.set_ylabel("GDDL Score", fontsize=14)
# Show grid
ax.grid(True)
# Adjust layout for better appearance
plt.tight_layout()
# Display the plot
plt.show()
---------------------------------------------------------------------------
FileNotFoundError Traceback (most recent call last)
<ipython-input-31-a47e650b06d9> in <cell line: 41>()
39
40 # Load RMSD and energy data
---> 41 rmsd_results = energy_array = np.array([float(line.split()[3].strip('\n')) for line in Path('rmsd.info').read_text().splitlines() if not line.startswith('!') and not line.startswith('Bank')], dtype=np.float64)
42 energy_array = np.array([float(line.split()[1]) for line in Path('GalaxyDock_fb.E.info').read_text().splitlines() if not line.startswith('!') and not line.startswith('Bank')], dtype=np.float64)
43
/usr/lib/python3.10/pathlib.py in read_text(self, encoding, errors)
1132 """
1133 encoding = io.text_encoding(encoding)
-> 1134 with self.open(mode='r', encoding=encoding, errors=errors) as f:
1135 return f.read()
1136
/usr/lib/python3.10/pathlib.py in open(self, mode, buffering, encoding, errors, newline)
1117 if "b" not in mode:
1118 encoding = io.text_encoding(encoding)
-> 1119 return self._accessor.open(self, mode, buffering, encoding, errors,
1120 newline)
1121
FileNotFoundError: [Errno 2] No such file or directory: 'rmsd.info'
The process of converting MOL2 files into SDF files for prolif processing.
[ ]:
#@title Converting docking results into SDF files
import subprocess as sp
from openbabel import pybel
mol2_file_path = f"/content/iitp_demonstrate/{target}/GalaxyDock_fb.mol2"
sdf_output_file = f"/content/iitp_demonstrate/{target}/GalaxyDock_fb_combined.sdf"
rank_mol2_file = f"/content/iitp_demonstrate/{target}/final_ligand.mol2"
sdf_rank_file = f"/content/iitp_demonstrate/{target}/final_ligand.sdf"
try:
with open(sdf_output_file, "w") as sdf_file:
for mol in pybel.readfile("mol2", mol2_file_path):
sdf_file.write(mol.write("sdf"))
print(f"Successfully combined all molecules of output files into {sdf_output_file}")
with open(sdf_rank_file, "w") as sdf_file:
for mol in pybel.readfile("mol2", rank_mol2_file):
sdf_file.write(mol.write("sdf"))
print(f"Successfully combined the selected rank molecule file into {sdf_rank_file}")
except Exception as e:
print(f"Error during .mol2 to .sdf conversion: {e}")
Successfully combined all molecules of output files into /content/iitp_demonstrate/2qeh/GalaxyDock_fb_combined.sdf
Successfully combined the selected rank molecule file into /content/iitp_demonstrate/2qeh/final_ligand.sdf
Utilizing Prolif, visualize the types of interactions between the ligand and protein in each protein-ligand docking pose from the GDDL output.
[ ]:
#@title Displaying protein-ligand interactions using ProLIF
import prolif as plf
from rdkit import Chem
poses_path = sdf_output_file
pose_iterable = plf.sdf_supplier(poses_path)
rdkit_prot = Chem.MolFromPDBFile(f'/content/iitp_demonstrate/{target}/{target}_pro.pdb', removeHs=False)
protein_mol = plf.Molecule(rdkit_prot)
fp = plf.Fingerprint()
fp.run_from_iterable(pose_iterable, protein_mol)
fp.plot_barcode(xlabel="Pose")
WARNING:MDAnalysis.coordinates.AMBER:netCDF4 is not available. Writing AMBER ncdf files will be slow.
/usr/local/lib/python3.10/dist-packages/MDAnalysis/topology/tables.py:52: DeprecationWarning: Deprecated in version 2.8.0
MDAnalysis.topology.tables has been moved to MDAnalysis.guesser.tables. This import point will be removed in MDAnalysis version 3.0.0
warnings.warn(wmsg, category=DeprecationWarning)
<Axes: xlabel='Pose'>
Visualize which residues of the protein interact with the ligand in the docking pose corresponding to the selected rank.
[ ]:
fp.plot_lignetwork(pose_iterable[rank])
Both the protein (PDB format) and ligand (mol2 format) files generated by GDDL can be downloaded.
[ ]:
#@title Saving docking results into Google Drive
from google.colab import files
output_pdb = 'pdb_processed.pdb'
files.download(output_pdb)
output_mol2 = 'GalaxyDock_fb.mol2'
files.download(output_mol2)
Section 4: Re-scoring GDDL docking results using BindingRMSD.¶
BindingRMSD CASF-2016 Docking power benchmark results
4-1. Re-scoring for Docked Protein-Ligand Complex¶
Performing BindingRMSD model using docking result file.
[ ]:
protein_file_path = f"/content/iitp_demonstrate/{target}/pdb_processed.pdb"
output_file = "/content/output.tsv"
model_path = "/content/BindingRMSD/save"
# Construct the command
command = [
"python3",
"/content/BindingRMSD/inference.py",
"-r", protein_file_path,
"-l", sdf_output_file,
"-o", output_file,
"--model_path", model_path
]
print( " ".join( command ))
sp.run(command, check=True)
# Run the command using subprocess
try:
sp.run(command, check=True)
print(f"Inference completed successfully. Results saved to {output_file}")
except sp.CalledProcessError as e:
print(f"Error during inference: {e}")
python3 /content/BindingRMSD/inference.py -r /content/iitp_demonstrate/2qeh/pdb_processed.pdb -l /content/iitp_demonstrate/2qeh/GalaxyDock_fb_combined.sdf -o /content/output.tsv --model_path /content/BindingRMSD/save
---------------------------------------------------------------------------
CalledProcessError Traceback (most recent call last)
<ipython-input-36-258f7d87d702> in <cell line: 16>()
14
15 print( " ".join( command ))
---> 16 sp.run(command, check=True)
17
18 # Run the command using subprocess
/usr/lib/python3.10/subprocess.py in run(input, capture_output, timeout, check, *popenargs, **kwargs)
524 retcode = process.poll()
525 if check and retcode:
--> 526 raise CalledProcessError(retcode, process.args,
527 output=stdout, stderr=stderr)
528 return CompletedProcess(process.args, retcode, stdout, stderr)
CalledProcessError: Command '['python3', '/content/BindingRMSD/inference.py', '-r', '/content/iitp_demonstrate/2qeh/pdb_processed.pdb', '-l', '/content/iitp_demonstrate/2qeh/GalaxyDock_fb_combined.sdf', '-o', '/content/output.tsv', '--model_path', '/content/BindingRMSD/save']' returned non-zero exit status 1.
The prediction results of Binding RMSD are saved as a TSV file, and the saved TSV file is displayed as a DataFrame using pandas.
The Predicted_RMSD column is generated using the Binding RMSD regression model, which directly predicts the RMSD of the bound ligand pose in a given protein-ligand complex. The Is_Above_2A column is generated using the Binding RMSD binary classification model, which predicts whether the RMSD of the bound ligand pose in a given protein-ligand complex is above 2 Å. For both models, values closer to 0 indicate better performance.
[ ]:
import pandas as pd
df = pd.read_csv( output_file, sep='\t')
df.head()
4-2. 3D Visualization: Crystal Structure vs Docked Poses¶
Re-score the poses of ligands docked using GDDL to a crystal protein-ligand complex using the Binding RMSD model. After sorting the results to select the best-predicted pose, visualize it in 3D using Py3Dmol.
Sort based on Predicted_RMSD and visualize the results.
[ ]:
#@title Visualizing crystal structure and docked poses
import py3Dmol
from rdkit import Chem
from rdkit.Chem import AllChem
import os
# Function to visualize PDB and specific ligand from SDF, along with a native ligand
def visualize_pdb_with_ligand_and_native(pdb_file, sdf_file, ligand_index, native_pdb_file=None):
# Load PDB structure
with open(pdb_file, 'r') as file:
pdb_data = file.read()
# Load SDF file and select the ligand by index
suppl = Chem.SDMolSupplier(sdf_file, sanitize=False)
if ligand_index < len(suppl) and suppl[ligand_index] is not None:
ligand = suppl[ligand_index]
ligand_block = Chem.MolToMolBlock(ligand) # Convert ligand to MOL block
else:
print(f"Invalid index: {ligand_index}. The file contains {len(suppl)} molecules.")
return
# Check if native ligand file exists
native_data = None
if native_pdb_file and os.path.isfile(native_pdb_file):
with open(native_pdb_file, 'r') as file:
native_data = file.read()
# Create 3D visualization
view = py3Dmol.view(width=800, height=600)
view.addModel(pdb_data, "pdb") # Load PDB file
view.setStyle({'cartoon': {'color': 'spectrum'}}) # Cartoon style for protein
# Add ligand to the visualization
view.addModel(ligand_block, "mol") # Load MOL block for ligand
view.setStyle({'model': 1}, {'stick': {'colorscheme': 'cyanCarbon'}}) # Stick style for ligand
# Add native ligand if available
if native_data:
view.addModel(native_data, "pdb") # Load native PDB
view.setStyle({'model': 2}, {'stick': {'colorscheme': 'magentaCarbon'}}) # Stick style for native ligand
view.zoomTo() # Adjust the view to fit all structures
return view
# Example usage
df_sorted = df.sort_values(by='Predicted_RMSD', ascending=True)
best_pRMSD_index = int(df_sorted.index[0])
native_pdb_file = f"/content/iitp_demonstrate/{target}/{target}_lig.pdb"
view = visualize_pdb_with_ligand_and_native(protein_file_path, sdf_output_file, best_pRMSD_index, native_pdb_file=native_pdb_file)
if view:
view.show()
Sort based on Is_Above_2A and visualize the results.
[ ]:
# Example usage
df_sorted = df.sort_values(by='Is_Above_2A', ascending=True)
best_pRMSD_index = int(df_sorted.index[0])
native_pdb_file = f"/content/iitp_demonstrate/{target}/{target}_lig.pdb"
view = visualize_pdb_with_ligand_and_native(protein_file_path, sdf_output_file, best_pRMSD_index, native_pdb_file=native_pdb_file)
if view:
view.show()
4-3. Crystal Ligand Pose vs Docked Ligand Pose: RMSD and Prediction Comparison¶
Load the precomputed RMSD file and create an RMSD list.
[ ]:
with open(f'/content/iitp_demonstrate/{target}/rmsd.info') as fp:
rmsd_results = []
for line in fp:
if not line.startswith('!'):
rmsd_results.append(float(line.split()[3].strip('\n')))
A code that plots the results predicted by the regression and binary classification models against the actual RMSD of the docked pose from the crystal structure.
[ ]:
#@title Comparing actual vs. predicted RMSD values
import pandas as pd
import matplotlib.pyplot as plt
import numpy as np
from scipy.stats import linregress, kendalltau, spearmanr
from sklearn.metrics import mean_absolute_error, mean_squared_error
def add_trendline_and_metrics(ax, x, y, title):
slope, intercept, r_value, p_value, std_err = linregress(x, y)
trendline = np.array(x) * slope + intercept
ax.plot(x, trendline, color="g", linestyle="--", label=f"y = {slope:.2f}x + {intercept:.2f}")
tau, _ = kendalltau(x, y)
spearman_corr, _ = spearmanr(x, y)
mae = mean_absolute_error(y, np.array(x) * slope + intercept)
rmse = np.sqrt(mean_squared_error(y, np.array(x) * slope + intercept))
ax.text(
0.05, 0.95,
f"$R^2$ = {r_value**2:.2f}\n"
f"p = {p_value:.2e}\n"
f"Kendall Tau = {tau:.2f}\n"
f"Spearman = {spearman_corr:.2f}\n"
f"MAE = {mae:.2f}\n"
f"RMSE = {rmse:.2f}",
transform=ax.transAxes,
fontsize=10,
verticalalignment="top",
)
ax.legend(loc="upper left")
# Create side-by-side scatter plots
fig, ax = plt.subplots(1, 2, figsize=(12, 6), sharey=False)
ax[0].scatter(rmsd_results, df["Predicted_RMSD"], color='b', alpha=0.7, edgecolors='k')
add_trendline_and_metrics(ax[0], rmsd_results, df["Predicted_RMSD"], "x_data vs y_data_predicted")
ax[0].plot([0, max(df["Predicted_RMSD"])], [0, max(df["Predicted_RMSD"])], color="orange", linestyle="-", label="y = x")
ax[0].set_title("RMSD Results vs Predicted RMSD")
ax[0].set_xlabel("RMSD Results")
ax[0].set_ylabel("Predicted RMSD")
ax[0].grid(True)
ax[0].legend()
ax[1].scatter(rmsd_results, df["Is_Above_2A"], color='r', alpha=0.7, edgecolors='k')
add_trendline_and_metrics(ax[1], rmsd_results, df["Is_Above_2A"], "x_data vs y_data_is_above_2a")
ax[1].plot([0, 2, 2, max(df["Predicted_RMSD"])], [0, 0, 1, 1], color="orange", linestyle="-", label="Threshold at 2")
ax[1].set_title("RMSD Results vs Is Above 2Å")
ax[1].set_xlabel("RMSD Results")
ax[1].set_ylabel("Probability (Is Above 2Å)")
ax[1].set_ylim(-0.05, 1.05)
ax[1].grid(True)
ax[1].legend()
plt.tight_layout()
plt.show()
[ ]: